Beruflich Dokumente
Kultur Dokumente
Relative Orientation of Close-Packed:, 8-Pleated Sheets in Proteins
Hochgeladen von
Parijat BanerjeeOriginaltitel
Copyright
Verfügbare Formate
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenCopyright:
Verfügbare Formate
Relative Orientation of Close-Packed:, 8-Pleated Sheets in Proteins
Hochgeladen von
Parijat BanerjeeCopyright:
Verfügbare Formate
Proc. Nati Acad. Sci.
USA
Vol. 78, No. 7, pp. 4146-4150, July 1981 Biochemistry
Relative orientation of close-packed ,8-pleated sheets in proteins
(twisted ,3sheets/protein secondary and tertiary structure)
CYRUS CHOTHIA*t AND JOEL JANINt
*William Ramsay, Ralph Foster and Christopher Ingold Laboratories, University College London, Gower Street, London WC1E 6BT, England; tMedical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, England; and *Unit6 de Biochimie Cellulaire, Department de Biochimie et Gen~tique Moleculaire, Institut Pasteur, 28 rue du Docteur Roux, 75724 Paris Cedex 15, France
Communicated by Max F. Perutz, March 30, 1981
ABSTRACT When (3-pleated sheets pack face to face in proteins, the angle between the strand directions of the two (3-sheets is observed to be near -30. We propose a simple model for f3sheet-to-(-sheet packing in concanavalin A, plastocyanin, y-crystallin, superoxide dismutase, prealbumin, and the immunoglobin fragment VREI. This model shows how the observed relative orientation of two packed (3-sheets is a consequence of (i) the rows of side chains at the interface being approximately aligned and (ii) the (3-sheet having a right-handed twist. The special amino acid composition ofresidues at the (-sheet-to-(3-sheet interfaces makes the contact surfaces essentially smooth and hydrophobic.
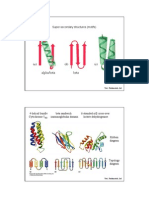
The three-dimensional structure of most globular proteins can be described as an assembly of close-packed a-helices and (& pleated sheets. It is therefore important to understand the rules that govern the packing of secondary structure (1-3). We have previously analyzed the packing ofa-helices on a-helices (4) and on (3-sheets (5). In this paper, we concentrate on (3sheet-to-,(sheet packing in a family of proteins that are formed by two (3pleated sheets packed face to face. Concanavalin A (6), plastocyanin (7), 'y-crystallin (8), superoxide dismutase (9), prealbumin (10), and the immunoglobulin domains (11) are representative members of this family, which also contains the coat proteins of tomato bushy stunt virus (12) and of southern bean mosaic virus (13), azurin (14), and other proteins. All these proteins have two (8-pleated sheets packed face to face and oriented so that the direction of the strands in one sheet is at an angle ofabout -300 to that in the other sheet, the minus sign indicating a left-handed screw rotation (Fig. 1). We show here that this characteristic orientation is a natural consequence of the right-handed twist of normal (pleated sheets and allows the close packing of side chains between the two (3sheets in contact. In other proteins formed by (3pleated sheets, such as trypsin or staphylococcal nuclease, the (3-pleated sheets are packed with the strand directions making an angle of about 900 (1). The principles applying to these structures are different and will be discussed elsewhere. A model for pacldng twisted (3-pleated sheets The P-pleated sheets observed in protein structures are not flat but twisted in a right-handed direction (16). In this section we describe the effects of introducing this twist into an idealized model of two (3-sheets packed face to face (Fig. 2). This model will be useful in understanding the structural descriptions presented in later sections. Fig. 2a illustrates one effect of the right-handed twist on a single polypeptide chain. On twisting ofthe main chain, the side chains that point up curve in the opposite direction to those that point down. As one looks down the chain, those that point up
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" in accordance with 18 U. S. C. 1734 solely to indicate this fact.
4146
curve from left to right, while those that point down curve from right to left. Fig. 2 b and c illustrates some of the effects of twisting two ,(3pleated sheets that are packed together. We show, on the left ofthe figures, two hypothetical untwisted (3-sheets packed with their rows of side chains aligned: those in the top (-sheet are parallel to those in the bottom. On the right of the figures we show what happens when these packed (3-sheets are twisted about an axis that is in the plane of their interface. We see that for the twisted close-packed (3-sheets: (i) The ends of the side chains that form contacts across the interface maintain their alignment. (ii) The main chains of the two sheets now point in
different directions. As one looks down the twist axis, the main chains of the top ,(3sheet go from left to right, while those of the bottom go from right to left (Fig. 2b). As one looks from the top (Fig. 2c), the main chains make an angle, which is negative because it represents a left-handed screw rotation. This angle, fQ, would be -350 if the top and bottom main chains are 10 A apart and the twist along the chains is 40/A. This angle of -35 would be increased or decreased by the rows of residues being not exactly aligned, by there being a different amount of twist, or both. If the side chains are not exactly aligned, and if the direction of the twist axis is closer to the direction of the strands in one (-sheet than to that in the other, the extent of the twist in the two (3-sheets will differ somewhat. Viewed from the end, the twisted (3-pleated sheets in projection take a cylindrical shape that Fig. 2b shows clearly. Note that the "barrel effect" (17) is observed even in the absence of a continuous network of hydrogen bonds joining the two (3sheets into a single one. In our model, top and bottom strands have their side chains pointing to each other and their main chains cannot hydrogen bond. The actual shape of the structure is not a "barrel" or a cylinder, but a twisted prism with a rectangular cross-section that rotates around the twist axis. The model described here supersedes a previous model of Chothia et al. (1). In the remainder ofthe paper, we shall discuss its relevance to real protein structures.
,B-Sheet-to-p-sheet packing in six proteins We used computer graphics to examine the detailed atomic structures of the six proteins or protein subunits illustrated in Fig. 1: concanavalin A, plastocyanin, y-crystallin domain I, superoxide dismutase, prealbumin, and immunoglobulin fragment VREI. For each protein, we determined the twist and the relative orientation of the (B-sheets and we analyzed the packing of amino acid residues at the (3-sheet to (3-sheet interfaces. P-Sheet Twist. The twist is conveniently expressed by the dihedral angle between pairs of adjacent strands. This angle is negative for all but three of 44 pairs of strands in the twelve (& sheets, indicating a normal right-handed twist of the strands,
Biochemistry: Chothia and janin
Proc. Natl. Acad. Sci. USA 78 (1981)
4147
2S
1472
2 &
747
243~~~S
042A
38
57
PIG
e-Cry
S3
Sod
Pro
VREI
FIG. 1. The 3-pleated sheets in concanavalin A (Con A), plastocyanin (Pla), y-crystallin (-,Cry), superoxide dismutase (Sod), prealbumin (Pra), and the immunoglobulin fragment VREI. *, Ca atoms in the "top" -3-sheet; 0, Ca atoms in the "bottom" 3sheet. Atomic coordinates are from the Cambridge Protein Data Bank (15), except for plastocyanin and 'tycrystallin, gifts of J. M. Guss, H. C. Freeman, and T. Blundell (7, 8).
which leads to a left-handed twist of the 3-sheets viewed from the edge. The mean of the distribution (Fig. 3) is -17 and the standard deviation is 9. The distribution is not significantly different from that found in a/P proteins, in which the mean dihedral angle between adjacent strands is - 19 with a standard deviation of 70 (4). The dihedral angle between the right-most and left-most strands of each (3-sheet is a measure of its total twist. It varies between -30 over six strands in the top (-sheet of concanavalin A and -65 over four strands for the top (3-sheet of prealbumin (Table 1). Thus, despite the rather narrow distribution in Fig. 3, the (3-sheets can differ widely in their total twist. Relative Orientation of the (Sheets. Relative orientation is fixed by the 0 angle of Fig. 2c. The values found for the six proteins are listed in Table 1. The values are in the range -30 + 150. Residue Packing at the 3-Sheet-to-p-Sheet Interface. Packing of atoms inside a protein structure may be studied by drawing sections through a space-filling model of the protein, generated by a computer from the atomic coordinates. We show such sections in Fig. 4, for the immunoglobulin fragment VREI and for the prealbumin subunit. The sections are cut perpendicular to the mean direction of the strands, so that the viewpoint is the same as in Fig. 2b, which makes comparison with the model easier. The sections in Fig. 4a are made through the front part of the interface, the sections in Fig. 4b, through the
middle, and the sections in Fig. 4c, through the back. A complete representation of the structures would involve stacking a number of such sections on top of each other. We see in Fig. 4 that: (i) The description of the prealbumin and VREI structures as made of two ,(3pleated sheets packed face to face is essentially correct. Residues not belonging to the (3 sheets are marked in dotted lines. They make little contribution to the internal packing. (ii) The (3sheet-to-,3sheet interface is made up of side chains pointing directly to each other. In each section, a line may be drawn that separates one (3-sheet from the other. Side chains from either side hardly cross this line. Thus, intercalation of side chain from top and bottom is the exception rather than the rule in these interfaces. In the six sections shown, it occurs once with Trp-35 in VREI (Fig. 4b), which fits its side chain into a hole in the bottom (3-sheet. Yet, the interface is not a plane. Due to the twist, the dividing line between the 3-sheets rotates clockwise from the front to middle to back sections. (iii) The rows of side chains are approximately aligned even though the main chains are not. Thus, we can follow the contacts between residues 12, 14, and 16 from strand 11-19 in the prealbumin bottom (8-sheet with residues 69, 71, and 73 from strand 67-74 of the top 3-sheet. Fig. 1 shows that the main chains of these two strands are at an angle of -30 or so. In Vmn residues 75, 73, and 71, from the bottom (&sheet strand 69-76, are in contact with residues 86, 35, and 33 from the two adjacent strands, 84-91 and 32-38, in the top (3-sheet and the angle between the main chains in the top and bottom
4148
Biochemistry: Chothia and Janin
STRAIGHT CHAINS TWISTED CHAINS
Proc. Natl. Acad. Sci. USA 78 (1981)
10
1OF
0
\+1
CL
0.
510.
(a) Single chain
w .0
E z
nFl-F
.
AL .
TFH
0 +10
m m
4~
I
I
W
I
-50 -40 -30 -20 -10 Dihedral angle
I
(b) Two packed sheets: end view
FIG. 3. Histogram of the twist angle (in degrees) between adjacent strands. The dihedral angle between adjacent strands in a 1-sheet expresses the twist of the 1-sheet. Ten values out of 44 are outside the range -25 to -5. They all involve strands at the edge of their respective 1-sheets.
(c) Two packed sheets: top view
FIG. 2. A model for the packing of 1pleated sheets. The main chain is represented by one small filled circle per residue, the side chains being large open circles. In a we show a single strand, untwisted or twisted in the normal right-handed direction. In b and c we show views of packed 13-sheets with two strands each. Only the side chains pointing into the interface are drawn.
/3sheets is again -300. The contact between the side chains is maintained over more than 15 A through the twist of the /3sheets, in accordance with the model described in Fig. 2. Sections cut through concanavalin A, y-crystallin, plastocyanin, and superoxide dismutase are similar in their general appearance to those shown here for prealbumin and the VREI fragment. The dihedral angle made by the main chain of pairs of strands in contact across the interface varies from - 100 to -55 around a mean value of -33. The variation results in part from the variable twist of the strands, and in part from an imperfect alignment of the rows of side chains. The largest misalignment occurs in plastocyanin and is responsible for its small value. Packing at the Edges of the 13-Sheets. All but 4 of the 24 strands that form the edges of the 12 /-sheets pack in a normal way, that is, like internal strands, even though their main chain is hydrogen bonded on one side instead of two. The four exceptions are of two types: (i) Residues 13-21 in plastocyanin and residues 3-13 in VREI form two sections of /-strand each, joined by a non-/-strand residue. The first section is part of the top /3-sheet, the second, part of the bottom /3-sheet (see Fig. 1). Though strand 13-21 in plastocyanin, or 3-13 in VREI, cannot pack as a whole in accordance with the model, each of the two sections does. This can be seen in Fig. 4 for VREI. (ii) In the
superoxide dismutase subunit, the edge strands 4-9 and 142148 hydrogen bond to each other: Val-5 to Gly-148 and Lys-9 to Cys-144. Residues 9 and 148 are at the corners of their sheets (Fig. 1) and local sharp twists in the /3-sheets turn their peptides so they are in a position to hydrogen bond to residues 144 and 5, respectively. Amino acid composition of the 13-pleated sheets and of their contact residues The 3pleated sheets of concanavalin A, plastocyanin, prealbumin, superoxide dismutase, and the VREI fragment contain a total of 355 residues, ofwhich 119 are involved in the /3-sheetto-/3-sheet interfaces through their side chain. The proportion of contact residues, 33%, is less than the 50% expected from a model in which half of the side chains in the /3-sheets point into the interface and half away from it. The discrepancy results in part from the /3-sheets being ofunequal size and not overlapping exactly, and in part from non-,&sheet residues packing between the ends of the /3-sheets. Table 2 gives the amino acid composition ofthe /-sheets and that ofthe contact residues in the /3-sheet-to-/3sheet interfaces.
/3-
Table 1. Twist and relative orientation of the ,&pleated sheets Relative orientation twist, ,t3 Sheet twistof the,13 13-Shee Protein Bottom sheets fl, Top Concanavalin A 30 (6) 80 (7) -30 40 (3)* 45 (3)* -25 y-Crystallin 40 (3)* 45 (3)* -20 Plastocyanin Prealbumin 65 (4) 45 (4) -35 55 (4) 35 (4) -45 Superoxide dismutase 60 (5)* 40 (4) -30 VREI The angles were calculated by fitting vectors to the main chain atoms of the strands. Previously we only fitted the Ca atoms (1) and in some cases the different fits give slightly different values for the angles. The top 13-sheet is that nearer to the reader in Fig. 1 and the bottom 1-sheet the one farther away. The values are given as O' (n), in which Ois the angle between n strands. The strands are those at the edges of the 1-sheets except in the cases marked *, for which we have excluded strands of three residues that have an unusually large or small twist and would therefore create a false impression of the extent of the overall 1sheet twist.
Biochemistry: Chothia and janin
Proc. Natl. Acad. Sci. USA 78 (1981)
4149
Immunoglobin fragment VREI
x=oA
a
-
X=+8A
mean a-sheet to P-sheet interface
\ vY /
V\1\/\/
cA b1
.
+x
7AA
X=-8A
x=oA
Prealbumin
X=+7A
FIG. 4. The (3-sheet-to-,B3sheet residue packing in prealbumin and VREI. Space-filling models of the proteins were generated in the computer. Sections were cut perpendicular to the mean direction of the strands in the (3-sheets-see the small schematicInset. Broken lines represent the edges of the van der Waals envelopes of the residues in one ,/3sheet and full lines the residues in the other (-sheet. Bar lines mark their mean interface. Dotted lines represent residues not in either (-sheet. Three sections separated by 1 A are superimposed, so that each picture describes a 2-A-thick slice. The x value gives the relative position of the middle section. The viewpoint of the proteins is the same as that in Fig. 2b and the two should be compared. The sections usually cut through two residues from each strand, one pointing into the interface and one away. Strand affiliation of the residues can be seen in Fig. 1.
The composition of the /3-sheets in our sample is not significantly different from average, taking the much larger sample of (3-sheets of Levitt (18) as a reference. Yet, the amino acid composition of the contact residues is unusual, with four amino acids, Leu, Val, Ile, and Phe, forming 61% of the total. This proportion is much greater than in the average /3-sheet, where it is 31%, or in the average protein interior, where it is 38% (19). The branched side chains of Val, Ile, and Leu make the surface of the /3-sheets approximately smooth (20). The most common configuration of aromatic residues has the side chain lying flat over the main chain. As a consequence, there is no regular intercalation of side chains in the /3-sheet-to-/3-sheet interfaces, a point that Fig. 4 illustrates. Like a-helix-to-,-sheet interfaces (5), but unlike a-helix-to-a-helix interfaces, in which intercalation of side chains is the rule (4), /3-sheet-to-/-sheet interfaces tend to be formed by the assembly of essentially smooth surfaces. /3Sheet residues forming contact with a-helices in a sample of eight a/,/ proteins (5) consist of 10% Leu, 24% Val, 14% Ile, and 6% Phe, which makes them similar to the residues involved
in the /sheet-to-/-sheet contacts studied here. Most probably, the large excess of branched hydrophobic residues in a-helixto-/3-sheet and in /3-sheet-to-,/3sheet contacts reflects the need to produce a well-packed interface. As a result of a-helices packing on both sides of the /sheets in a//3 proteins, 67% of the /3-sheet residues are involved in the interfaces, against 33% here. The influence of the contact residues on the amino acid composition of their /sheets is therefore more evident. Lifson and Sander (20, 21) have noted that parallel and antiparallel /3 pleated sheets differ in their amino acid compositions, and especially that the former are enriched in Val and Ile. We do not think that this indicates a preference of these types of residues for the parallel association of strands, but that parallel /sheets are found almost exclusively in a//3 proteins, where the a-helices make the necessary connections between the strands, whereas all 83-proteins have mostly antiparallel strands (22, 23). Conclusion In this paper we have described how, in the class of protein structures illustrated in Fig. 1, 83-pleated sheets pack together. Other publications have been concerned with their strand to-
4150
Biochemistry: Chothia and Janin
Troc. Natt Acad., Sci.. USA 78 (1981)
Table 2. Amino acid composition of the, (-sheets and of the (sheet-to-3-sheet contact residues in five proteins Composition, mol % Contact residues in Average PSheets in the five proteins the five proteins .13-sheet Residue 12 17 11.9 Val 16 8 Leu 7T0 15 7.8 7 Ile 13 6 4.6 Phe 10 7.6 8 Ala 9 6 Thr 7.7 5 Tyr 4.8 6 7 4 8.7 Gly 1 1.6 3 Cys 1 3 1.5 Met 3 2 2.5 GIn 2 2 1.7 Trp 5 1 5.4 Lys 4 1 4.3 Asp 4 1 3.8 Glu 1 3.3 3 Asn 0 8 Ser 7.6 0 2 3.0 Arg 0 4 2.6 His 0 1 2.5 Pro
101 100 Total 9.9 The percentage amino acid composition of the 355 3-sheet residues in concanavalin A, plastocyanin, prealbumin, superoxide dismutase, and VREI is compared with (i) the 1555 residues representing an average (3-sheet (18) and (ii) the 119 residues forming(-sheet-to-(-sheet contacts in the five proteins.
ofthe regions involved in the association. On these smooth surfaces a-helices with their axis parallel to the strands, or a ,( sheet turned by -300, form a complementary surface that fits with little need for rearrangement of the side chains at the interface (24, 25). The association is stabilized by the removal of hydrophobic surfaces from contact with the solvent, which yields a favorable release of hydrophobic free energy.
We are grateful to Dr. Peter Pauling for encouragement, John Cresswell for the illustrations, and Dr. Michael Levitt and Douglas Richardson for computer programs. We thank Drs. J. M. Guss, H. C. Freeman, S. C. Harrison, E. T. Adman, and T. Blundell for the atomic coordinates oftheir protein structures prior to publication. C.C. is an E.P.A. Cephalosporin Fund Senior Research Fellow ofthe Royal Society. This work was supported by grantsto Dr. Peter Pauling from the2U.S. National Institute of General Medical Sciences (1-ROl-GM25435). and from the Science Research Council.
1. Chothia, C., Levitt, M. & Richardson, D. (1977) Proc. Natl Acad. Sci. USA 74, 4130-4134. 2. Richmond, T. J. & Richards, F. M. (1978)J. Mol Biol 119, 537555. 3. Cohen, F. E., Steinberg, M. J. E. &.Taylor, W. R. (1980) Nature (London) 285, 378-382. 4. Chothia, C., Levitt, M.,& Richardson, D. (1981)J. MoL BioL 145, 215-250. 5. Janin, J. & Chothia, C. (1980) J. Mol, Biol. 143, 95-128. 6. Reeke, G. N., Becker, J. W. & Edelman, G. M. (1975)J. Biol Chem. 250, 1525-1547. 7. Colman, P. M., Freeman, H. C., Guss, J. M., Murata, M., Norris, V. A. & Ramshaw, J. A. M. (1978) Nature (London) 272, 319324. 8. Blundell, T., Lindley, P., Miller, L., Moss, D., Slingsby, C., Tickle, I., Turnell, B. & Wistow, G. (1981) Nature (London) 289, 771-777. 9. Richardson, J. S., Thomas, K. A., Rubin, B. H. & Richardson, D. C. (1975) Proc. Natl Acad. Sci. USA 72, 1349-1353. 10. Blake, C. C. F., Geisow, M. J., Oatley, S. J., Rerat, B. &-Rerat, C. (1978)J. Mol Biol 121, 339-356. 11. Epp, O., Colman, P., Fehlhammer, H., Bode, W., Schiffer, M., Huber, R. & Palm, W. (1974) Eur. J. Biochem. 45, 513-524. 12. Harrison, S. C., Olson, A. J., Schutt, C. E., Winkler, F. K. & Bricogne, G. (1978) Nature (London) 276, 368-373. 13. Abad-Zapatero, C., Absel-Meguid, S. S., Johnson, J. E., Leslie, A. G. W., Rayment, I., Rossmann, M. G., Suck, D. & Tsukihara, T. (1980) Nature (London) 286, 33-39. 14. Adman, E. T., Stenkamp, R. E., Sieker, L. C. & Jensen, L. H. (1978)J. Mol Biol 123, 35-48. 15. Bernstein, F. C., Koetzle, T. F., Williams, G. J. B., Meyer, E. F., Jr., Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977)J. Mol Biol 112, 535-542. 16. Chothia, C. (1973) J. Mol Biol 75, 295-302. 17. Richardson, J. S., Richardson, D. C., Thomas, K. A., Silverton, E. W. & Davies, D. R. (1976)J. Mol Biol 102, 221-235. 18. Levitt, M. (1978) Biochemistry 17, 4277-4384. 19. Janin, J. (1979) Nature (London) 277, 491-492. 20. Lifson, S. & Sander, C. (1980),in Protein Folding, ed. Jaenicke, R. (Elsevier, Amsterdam), pp. 289-316. 21. Lifson, S. & Sander, C. (1979) Nature (London) 282,109-111. 22. Levitt, M. & Chothia, C. (1976) Nature (London) 261, 552-558. 23. Richardson, J. S. (1977) Nature (London) 268, 495-500. 24. Janin, J., Wodak, S.,; Levitt, M. & Maigret, B. (1978)J. Mol Biol 125, 357-386. 25. Gelin, B. R. & Karplus, M. (1979) Biochemistry 18, 1256-1268.
pology (22-24) and with the patterns formed by their contact residues (3). Taken together, these papers give us a clear picture of the general principles that govern the three-dimensional structure of these proteins. The reason for the rows of side chains being approximately aligned at the interface is that it allows the (3-sheets to be twisted and have a large contact surface. If two twisted (3-sheets are put face to face at a large value of fi they splay apart. This occurs in the other class of structures formed by (-sheets in which, f
900. The packing principles that determine these structures discussed elsewhere. The forces that determine the three-dimensional structures of proteins are too complex to be described in exact detail. Yet, the elaborate architecture of proteins obeys rules that can be revealed by an empirical analysis of available structural data. We have attempted to draw such rules for the assembly. of proteins made of a-helices packed on a-helices (4), or on P-sheets (5), and now, for proteins made of two 3-sheets packed face to face. In all three sorts of structures, the basic geometry of the association is fixed by that of the elements. Thus, the righthanded twist of(,&pleated sheets is the prominent feature.in ahelix-to-,&sheet and (sheet-to-(-sheet association. The (sheets appear as essentially smooth twisted surfaces, their smoothness resulting from a restricted amino acid composition
are
Das könnte Ihnen auch gefallen
- Mls 014 Hematology 2 Lab Quiz 3Dokument11 SeitenMls 014 Hematology 2 Lab Quiz 3CliffordNoch keine Bewertungen
- Hekix SheetDokument5 SeitenHekix SheetParijat BanerjeeNoch keine Bewertungen
- Secondary Structure - Beta StrandsDokument2 SeitenSecondary Structure - Beta StrandsAatmaanandaNoch keine Bewertungen
- Journal: Structure of SpinelDokument14 SeitenJournal: Structure of SpinelRaul EvertonNoch keine Bewertungen
- Structure of SpinelDokument14 SeitenStructure of SpinelAlin DrucNoch keine Bewertungen
- Sundance Bilson-Thompson, Jonathan Hackett and Louis H. Kau Man - Particle Topology, Braids and Braided BeltsDokument21 SeitenSundance Bilson-Thompson, Jonathan Hackett and Louis H. Kau Man - Particle Topology, Braids and Braided BeltsMopadDeluxeNoch keine Bewertungen
- Dynamic Monte Carlo Study of The Folding of A Six-Stranded Greek Key Globular ProteinDokument5 SeitenDynamic Monte Carlo Study of The Folding of A Six-Stranded Greek Key Globular ProteinMercurio157Noch keine Bewertungen
- James H. White and Nicholas R. Cozzarelli - A Simple Topological Method For Describing Stereoisomers of DNA Catenanes and KnotsDokument5 SeitenJames H. White and Nicholas R. Cozzarelli - A Simple Topological Method For Describing Stereoisomers of DNA Catenanes and KnotsLokosooNoch keine Bewertungen
- Yidun Wan - Effective Theory of Braid Excitations of Quantum Geometry in Terms of Feynman DiagramsDokument23 SeitenYidun Wan - Effective Theory of Braid Excitations of Quantum Geometry in Terms of Feynman DiagramsMopadDeluxeNoch keine Bewertungen
- Charles W. Trigg - Collapsible Models of The Regular OctahedronDokument5 SeitenCharles W. Trigg - Collapsible Models of The Regular OctahedronEduardo MullerNoch keine Bewertungen
- 2nd RefDokument12 Seiten2nd RefRwitajit MajumdarNoch keine Bewertungen
- J.A. Tuszynski, J.A. Brown, E. Crawford, E.J. Carpenter, M.L.A. Nip, J.M. Dixon and M.V. Sataric: Molecular Dynamics Simulations of Tubulin Structure and Calculations of Electrostatic Properties of MicrotubulesDokument16 SeitenJ.A. Tuszynski, J.A. Brown, E. Crawford, E.J. Carpenter, M.L.A. Nip, J.M. Dixon and M.V. Sataric: Molecular Dynamics Simulations of Tubulin Structure and Calculations of Electrostatic Properties of MicrotubulesUloffNoch keine Bewertungen
- Chapter 6. QuasicristalesDokument48 SeitenChapter 6. QuasicristalesafnaftanelNoch keine Bewertungen
- Spiro CompoundDokument11 SeitenSpiro CompoundGanesamoorthy ThirunarayananNoch keine Bewertungen
- Basic Concepts of Crystalline StructureDokument60 SeitenBasic Concepts of Crystalline StructureKhen Mehko Ojeda100% (1)
- Systematics of The Spinel Structure TypeDokument23 SeitenSystematics of The Spinel Structure TypeAlin DrucNoch keine Bewertungen
- Molecule-Based Materials The Structural Network Approach C8Dokument16 SeitenMolecule-Based Materials The Structural Network Approach C8BRUNO RAMOS DE LIMANoch keine Bewertungen
- Crystallography and The Penrose Pattern: London EnglandDokument5 SeitenCrystallography and The Penrose Pattern: London EnglandedirozemberghNoch keine Bewertungen
- Biochemistry LN05-1Dokument14 SeitenBiochemistry LN05-1Rahaf Al-muhtasebNoch keine Bewertungen
- Dot PlotDokument2 SeitenDot PlotdarshowlkatNoch keine Bewertungen
- All Of: B andDokument1 SeiteAll Of: B andKetanNoch keine Bewertungen
- OB (Oligonucleotide/oligosaccharide Binding) - Fold: Structural and Functional Solution For Non-HomologousDokument7 SeitenOB (Oligonucleotide/oligosaccharide Binding) - Fold: Structural and Functional Solution For Non-HomologousSamra KanwalNoch keine Bewertungen
- Dna StructureDokument7 SeitenDna StructurehoneyworksNoch keine Bewertungen
- A Tight-Binding Approach To Uniaxial Strain in GrapheneDokument9 SeitenA Tight-Binding Approach To Uniaxial Strain in GrapheneNathan Scott NicholsNoch keine Bewertungen
- Inflation Rules, Band Structure, and Localization of Electronic StatesDokument8 SeitenInflation Rules, Band Structure, and Localization of Electronic StatesEduardo SaavedraNoch keine Bewertungen
- ArticuloDokument3 SeitenArticuloJose Manuel OrtizNoch keine Bewertungen
- Fig. 1: Schematic Diagram and Cryoem Image Analysis of The Flagellar Basal BodyDokument3 SeitenFig. 1: Schematic Diagram and Cryoem Image Analysis of The Flagellar Basal BodyMariusNoch keine Bewertungen
- Molecular DinamysDokument16 SeitenMolecular DinamysJair Torres DuranNoch keine Bewertungen
- STUDY168@373458Dokument9 SeitenSTUDY168@373458riyazqureshi27707Noch keine Bewertungen
- 5 BackupDokument29 Seiten5 BackupsadiksnmNoch keine Bewertungen
- TrussesDokument28 SeitenTrussesAbdul GaniNoch keine Bewertungen
- Take Home Message From Lecture 5Dokument33 SeitenTake Home Message From Lecture 5Carlos Eduardo LinaresNoch keine Bewertungen
- A New Principle of RNA Folding Based On Pseudoknotting1985 Pleij CWDokument15 SeitenA New Principle of RNA Folding Based On Pseudoknotting1985 Pleij CWxprakashNoch keine Bewertungen
- Hydrophobically-Driven Self-Assembly: A Geometric Packing AnalysisDokument4 SeitenHydrophobically-Driven Self-Assembly: A Geometric Packing AnalysisemediageNoch keine Bewertungen
- Cong JMolBiol 2008Dokument6 SeitenCong JMolBiol 2008huouinkyoumaNoch keine Bewertungen
- The Crystal Structure: A U - STDokument23 SeitenThe Crystal Structure: A U - STMansyur SQeNoch keine Bewertungen
- Crystal Lattices: 2.1 The LatticeDokument10 SeitenCrystal Lattices: 2.1 The LatticeBridget GwenNoch keine Bewertungen
- Recovered PDF 7 PDFDokument6 SeitenRecovered PDF 7 PDFeid elsayedNoch keine Bewertungen
- Organic ChemistryDokument29 SeitenOrganic ChemistrySerhan ÜnverNoch keine Bewertungen
- Assignment Topics With Materials 1. What Are The Advanced Materials?Dokument39 SeitenAssignment Topics With Materials 1. What Are The Advanced Materials?joshua nithishNoch keine Bewertungen
- Dna 3dmodel PaperDokument2 SeitenDna 3dmodel PaperluismitlvNoch keine Bewertungen
- M06 MATH 0231 01 SM C06v2-0-2Dokument4 SeitenM06 MATH 0231 01 SM C06v2-0-2WT ANoch keine Bewertungen
- Isabel K. Darcy - Modeling protein-DNA Complexes With TanglesDokument19 SeitenIsabel K. Darcy - Modeling protein-DNA Complexes With TanglesYopghm698Noch keine Bewertungen
- ATP Synthase and Other Motor ProteinsDokument3 SeitenATP Synthase and Other Motor Proteinsmoviemac7Noch keine Bewertungen
- Build A Paper Model of DNADokument2 SeitenBuild A Paper Model of DNAAlan MontalvoNoch keine Bewertungen
- Extra Notes PolyphaseDokument32 SeitenExtra Notes PolyphaseSathia RajNoch keine Bewertungen
- CH 05Dokument31 SeitenCH 05AbuNoch keine Bewertungen
- NIH Public Access: Author ManuscriptDokument25 SeitenNIH Public Access: Author ManuscriptLokosooNoch keine Bewertungen
- An Amino Acid Code for Β-sheet Packing StructureDokument13 SeitenAn Amino Acid Code for Β-sheet Packing StructureVenkata Suryanarayana GorleNoch keine Bewertungen
- Part 5Dokument6 SeitenPart 5Mithra LakshmiNoch keine Bewertungen
- Actomyosin RingsDokument8 SeitenActomyosin RingsPrafull SharmaNoch keine Bewertungen
- Stabilization of Phage T4 by Disulfide Bonds: Iysozyme EngineeredDokument5 SeitenStabilization of Phage T4 by Disulfide Bonds: Iysozyme Engineeredsruthijacob123Noch keine Bewertungen
- Orris 1973Dokument20 SeitenOrris 1973Supantho ChaudhuriNoch keine Bewertungen
- Matrix-Fracture Transfer Shape Factors For Dual-Porosity Simulators (Lim & Aziz)Dokument10 SeitenMatrix-Fracture Transfer Shape Factors For Dual-Porosity Simulators (Lim & Aziz)consultariskaNoch keine Bewertungen
- A. Jákli Et Al - Evidence For Triclinic Symmetry in Smectic Liquid Crystals of Bent-Shape MoleculesDokument4 SeitenA. Jákli Et Al - Evidence For Triclinic Symmetry in Smectic Liquid Crystals of Bent-Shape MoleculesKonnasderNoch keine Bewertungen
- GarterDokument4 SeitenGarterapi-3733260Noch keine Bewertungen
- Fractal Dimensions of A Green Broccoli and A White CauliflowerDokument13 SeitenFractal Dimensions of A Green Broccoli and A White CauliflowerAriel TeranNoch keine Bewertungen
- Coulomb's Law ExperimentDokument12 SeitenCoulomb's Law Experimentchafraed67% (3)
- Reviews in Computational Chemistry, Volume 31Von EverandReviews in Computational Chemistry, Volume 31Abby L. ParrillNoch keine Bewertungen
- Amorphous Semiconductors: Structural, Optical, and Electronic PropertiesVon EverandAmorphous Semiconductors: Structural, Optical, and Electronic PropertiesNoch keine Bewertungen
- Review Article: Host Cell Signalling and Mechanisms of EvasionDokument15 SeitenReview Article: Host Cell Signalling and Mechanisms of EvasionParijat BanerjeeNoch keine Bewertungen
- Toxoplasma Gondii: Infection Specifically Increases The Levels of Key Host MicrornasDokument8 SeitenToxoplasma Gondii: Infection Specifically Increases The Levels of Key Host MicrornasParijat BanerjeeNoch keine Bewertungen
- MicroDokument11 SeitenMicroParijat BanerjeeNoch keine Bewertungen
- Analysis of Microrna Transcriptome by Deep Sequencing of Small Rna Libraries of Peripheral BloodDokument18 SeitenAnalysis of Microrna Transcriptome by Deep Sequencing of Small Rna Libraries of Peripheral BloodParijat BanerjeeNoch keine Bewertungen
- Solvatochromic EffectDokument17 SeitenSolvatochromic EffectParijat Banerjee100% (1)
- Molecule of The Month: Molecular-Chameleon: Solvatochromism at Its Iridescent Best!Dokument4 SeitenMolecule of The Month: Molecular-Chameleon: Solvatochromism at Its Iridescent Best!Parijat Banerjee100% (1)
- Factors Regulating The Transcription of Eukaryotic Protein Coding Genes and Their Mechanism of Action-A ReviewDokument14 SeitenFactors Regulating The Transcription of Eukaryotic Protein Coding Genes and Their Mechanism of Action-A ReviewParijat BanerjeeNoch keine Bewertungen
- Molecular BiologyDokument13 SeitenMolecular BiologyParijat BanerjeeNoch keine Bewertungen
- Geszvain LandickDokument12 SeitenGeszvain LandickParijat BanerjeeNoch keine Bewertungen
- Codon Adaptation IndexDokument9 SeitenCodon Adaptation IndexParijat BanerjeeNoch keine Bewertungen
- Mirkin DNA TopologyDokument11 SeitenMirkin DNA TopologyParijat BanerjeeNoch keine Bewertungen
- Stress ResponseDokument16 SeitenStress ResponseParijat BanerjeeNoch keine Bewertungen
- Transcription FactorsDokument32 SeitenTranscription FactorsParijat BanerjeeNoch keine Bewertungen
- Ch13c MotifsDokument6 SeitenCh13c MotifsParijat BanerjeeNoch keine Bewertungen
- Symmetry Operations 42Dokument43 SeitenSymmetry Operations 42Parijat BanerjeeNoch keine Bewertungen
- Techniques: Interpreting Chromatin Immunoprecipitation ExperimentsDokument5 SeitenTechniques: Interpreting Chromatin Immunoprecipitation ExperimentsParijat BanerjeeNoch keine Bewertungen
- Structural Analysis of Protein Structure: Circular Dicroism Fluorescence X-Ray NMRDokument75 SeitenStructural Analysis of Protein Structure: Circular Dicroism Fluorescence X-Ray NMRParijat BanerjeeNoch keine Bewertungen
- Vitamin K, D and A Bind To The SARS CoV 2 Spike Protein - Ray Peat ForumDokument12 SeitenVitamin K, D and A Bind To The SARS CoV 2 Spike Protein - Ray Peat ForumDarci HallNoch keine Bewertungen
- Protein: What Is ADokument2 SeitenProtein: What Is AFilip KesteliNoch keine Bewertungen
- Determination of Lipids-2Dokument9 SeitenDetermination of Lipids-2Paola Rivera DiazNoch keine Bewertungen
- Electron Transport ChainDokument1 SeiteElectron Transport ChainKeith Justine AbabaoNoch keine Bewertungen
- 2.5 Enzyme Skeleton NotesDokument8 Seiten2.5 Enzyme Skeleton Notesadri baigorriNoch keine Bewertungen
- Cy To SkeletonDokument10 SeitenCy To SkeletonWaqas KhanNoch keine Bewertungen
- Mechanisms of Coagulation and Fibrinolysis (Autosaved)Dokument60 SeitenMechanisms of Coagulation and Fibrinolysis (Autosaved)Tom Anthony TonguiaNoch keine Bewertungen
- Section XIV - Lipid TransportDokument2 SeitenSection XIV - Lipid TransportKatharine NervaNoch keine Bewertungen
- Amino Acids and Their Structure: Cooh-Ch-Nh2 - RDokument12 SeitenAmino Acids and Their Structure: Cooh-Ch-Nh2 - RRica NorcioNoch keine Bewertungen
- Iron-Sulfur Protein - WikipediaDokument21 SeitenIron-Sulfur Protein - Wikipediaamar hattimareNoch keine Bewertungen
- Proteins Ppt. (By CDM)Dokument31 SeitenProteins Ppt. (By CDM)Aldren MarianoNoch keine Bewertungen
- G-protein-Coupled ReceptorsDokument24 SeitenG-protein-Coupled ReceptorsNaimi Amalia hatimahNoch keine Bewertungen
- Fatty Acids + Physico-Chemical PropertiesDokument39 SeitenFatty Acids + Physico-Chemical PropertiesSaloni AryaNoch keine Bewertungen
- Levinthal's ParadoxDokument3 SeitenLevinthal's Paradoxperception888Noch keine Bewertungen
- Smooth and Granular Endoplasmic Reticulum RibosomesDokument26 SeitenSmooth and Granular Endoplasmic Reticulum RibosomesArfiNoch keine Bewertungen
- Lauric Oil Specification PDFDokument1 SeiteLauric Oil Specification PDFAndi crasherNoch keine Bewertungen
- ICH Q6B Service LeafletDokument2 SeitenICH Q6B Service LeafletM-ScanNoch keine Bewertungen
- Corn Gluten Meal - Tables of Composition and Nutritional Values of Feed Materials INRA CIRAD AFZDokument11 SeitenCorn Gluten Meal - Tables of Composition and Nutritional Values of Feed Materials INRA CIRAD AFZKatherine Leyla Anchayhua TorresNoch keine Bewertungen
- Normal Digestion / Absorption of FatDokument5 SeitenNormal Digestion / Absorption of FatMarc Michael Dela CruzNoch keine Bewertungen
- The Citric Acid CycleDokument29 SeitenThe Citric Acid CyclejaleemjamesNoch keine Bewertungen
- Protein-Protein InteractionDokument15 SeitenProtein-Protein InteractionMateNoch keine Bewertungen
- 1 6 Monomer Drawing PracticeDokument4 Seiten1 6 Monomer Drawing PracticeSiddharth RajendranNoch keine Bewertungen
- DownloadDokument4 SeitenDownloadMarie Constance Therese PacquingNoch keine Bewertungen
- World of The Cell 7th Edition Becker Test BankDokument17 SeitenWorld of The Cell 7th Edition Becker Test Bankthuygladys5x0100% (23)
- Bile Acid SequestrantsDokument4 SeitenBile Acid SequestrantsFebrian IsharyadiNoch keine Bewertungen
- Mayonnaise Power PointDokument13 SeitenMayonnaise Power PointArs SaraNoch keine Bewertungen
- Crystal - Res.ku - Edu Taksnotes Biol 638 Notes CHP 16Dokument19 SeitenCrystal - Res.ku - Edu Taksnotes Biol 638 Notes CHP 16shahjafferNoch keine Bewertungen
- Fed State of MetabolismDokument40 SeitenFed State of MetabolismBHARANIDHARAN M.VNoch keine Bewertungen
- Lipids and Dyslipoproteinemia - 2020Dokument116 SeitenLipids and Dyslipoproteinemia - 2020Aria Jean MostajoNoch keine Bewertungen