Beruflich Dokumente
Kultur Dokumente
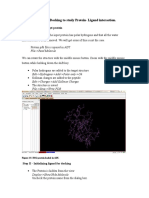
Autodock Tools Tutorial for Protein-Ligand Docking
Hochgeladen von
rednriOriginalbeschreibung:
Originaltitel
Copyright
Verfügbare Formate
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenCopyright:
Verfügbare Formate
Autodock Tools Tutorial for Protein-Ligand Docking
Hochgeladen von
rednriCopyright:
Verfügbare Formate
Tutorial for Autodock and Autodock Tools I. Establishing Access to the Programs A. Autodock is in /usr/local/Autodock.
The executables are autodock4 and autogrid4. Set up aliases in your .bashrc file: alias autodock3=/usr/local/Autodock/autodock4 alias autogrid3=/usr/local/Autodock/autogrid4 B. Autodock tools are in /usr/local/MGLTools-1.5.4. Add to your .bashrc:: alias pmv=/usr/local/MGLTools-1.5.4/bin/pmv alias adt=/usr/local/MGLTools-1.5.4/bin/adt source /usr/local/MGLTools-1.5.4/bin/mglenv.sh The aliases access the main parts of the tools; the last line sets up the Python environment for the tools. II. Preparing the Files A. Two files in Protein DataBank (pdb) format are required for a docking experiment: a structure for the protein and a structure for the ligand. B. In general, the protein structure will be downloaded from the Protein DataBank (www.rcsb.org), and the ligand structure will be created with one of our modeling applications. You should inspect both files using less or a text editor. In the file from the PDB you want to identify any non-protein species that will be removed later. This includes water (HOH) or other solvents, sugars, and so on. Make a list of the types and their abbreviations. Make sure you have a single copy of the protein in the file. Many proteins crystallize as dimers or trimers, with each copy of the protein containing a catalytic site. You must delete all extra chains. There are a few proteins the HIV protease is one in which the catalytic site is formed by the interaction of the chains. In this case, keep both chains. In the ligand file, check the column that lists the residue or other structure of which each atom is a part, and make sure that you have the three-letter code for the amino acids (if the ligand is a peptide), or a code of your choice for other ligands. PCModel puts UNK in this column when writing a pdb file. Change this to a three-letter acronym for your ligand so that the ligand can be selected properly for display in a program like JMol.
These files should be in the directory in which you choose to start. C. If you are working with the tutorial from the AutoDock web site, Using AutoDock with AutoDock Tools, the file as downloaded is both tarred and gzipped (extensions .tar.gz). this means that multiple files have been combined into one (tar) and compressed (gz). You must extract these by typing in a shell window tar zxvf filename.tar.gz (without the quotes, of course). This will create your two pdb files and a subdirectory containing the results from the docking for analysis. III. Editing the Protein PDB File with AutoDock Tools (ADT) We are going to fix any problems with the PDB files, such as missing bonds or atoms, and remove extraneous structures such as water molecules. Before beginning this section, inspect the PDB file to learn what such structures may be present. We want to keep only the protein and such cofactors as may be bound to it naturally. A. Start the ADT by typing adt in a shell window in the directory where you have placed your PDB files. Figure 1 showsthe ADT interface. The ADT window has several parts: (1) At the top are menus that access the functionality of PMV, the Python molecular viewer, including reading and writing coordinates, creating images, and modifying coordinates (2) A row of buttons for quick access the PMV functions (3) Menus to access AutoDock Tools functions. (4) The molecule viewer window (5) The Dashboard, for manipulating the molecule display. B. Load the protein From the File menu, choose Read Molecule, highlight the PDB file for your protein, and click Open. Or, right click on PMV Molecules at the bottom of the window and choose the protein pdb file. For the tutorial, select hsg1.pdb (1). As displayed, the protein appears white. You can color it according to any of several schemes by selecting Color from the main menu bar, and
choosing any of the schemes listed there. In the widget that pops up, click on All Geometries, and then on OK. You now have a pretty picture. Alternatively, click on the diamond under atom on the dashboard to color by atom type. (Figure 2)
Figure 1 The AutoDock Tools Window
C. Next, we remove the waters (those little red dots all over the display). On the main menu bar, choose Select, Select From String. A widget pops up (Figure 3): In the Residue entry box, type HOH*. The asterisk is a wild card, used in case the waters are numbered. It means accept any characters at
all following the H. Then type * in the Atom entry box; again the * is a wild card, this time meaning any atom. Click select. If the PCOM level is not already set to Atom (yellow button), you will get a message asking if setting it there is OK. Click Yes. Then click Dismiss. You should now see all of the waters marked with small yellow crosses (Figure 4).
Figure 2. Protein hsg1 Loaded and Colored
On the main menu bar, choose Edit, Delete, Delete AtomSet. ADT will ask if you really want to do this. Click Continue. The selected waters will disappear.
Repeat this process for any other small molecules you need to get rid of (check your pdb file!), such as other solvents like isopropyl alcohol (IPA), N-acetylglucosamine (NAG), and other relics of the isolation and crystallization procedures. Whole chains similarly can be deleted using the Chain entry box.
Figure 3 The Select from String Widget
D. X-ray crystallography usually does not locate hydrogens; hence most PDB files do not include them. But hydrogens, particularly those that can hydrogen bond, are important in binding ligands. (1) Choose Edit, Hydrogens, Add. Usually, you will choose Polar Only in the widget that pops up, using Method noBondOrder, and renumbering OK. This set of selections will add hydrogens to backbone N, and to amine and hydroxyl side side chains. You will see them appear. (2) Hide the protein by clicking the little gray box under Show/Hide. If you wanted to interrupt processing at this point, you could choose File -> Save -> Write PDB; however, we are going to add charges and AutoDock atom types.
IV. Preparing the Ligand File with ADT In this section information is added to the ligand pdb file to select bonds about which segments of the ligand will be rotated. Charges are added, using the Kollman scheme for peptide ligands and Gasteiger charges for other structures.
Before beginning, turn off display of the protein structure: click the Show/Hide Molecule button, and then click the button next to the name of your protein. Click the show/Hide button again to close the widget.
Figure 4 Water Molecules Selected for Deletion
A. Load the Ligand
(1) From the tan menubar just above the view window select: Ligand, Input Molecule, Open Molecule. Click the file types button on the widget that appears and select PDB files. Then choose the file containing your ligand, and click Open. In the tutorial, this is ind.pdb After a pause, a message will pop up, looking something like this (Figure 5) Figure 5. Summary of Actions on Ligand:
ADT has done its job of assigning charges, making sure all hydrogens are present, and detecting planar carbons in rings. TORSDOF is torsional degrees of freedom. B. Selecting Rotatable Segments The initial step is selecting a set of atoms with respect to which other groups of atoms will be rotated; this is called the Rigid Root. ADT tries to select a Root with the minimum number of rotatable branches. A central benzene ring is a popular choice. (1) On the tan menubar, select Ligand -> Torsion Tree -> Detect Root. A small green dot will appear, marking the choice. (2) Next, select Ligand -> Torsion Tree -> Choose Torsions. The Torsion Count widget appears (Figure 6).
The widget offers you several general selections of rotatable and non-rotatable bonds. (a) making peptide backbone bonds non-rotatable locks in the secondary structure of the peptide; normally this is a poor choice
(b) At laboratory temperatures, the barrier to rotation around the CO-N bond of an amide is too high to permit free rotation; if the ligand has amide functions, click this button.
Figure 6. The Torsion Count Widget
Rotatable bonds also may be selected individually. If you do this, be careful! Bonds within ring MUST be non-rotatable, and no more than 32 rotatable bonds may be selected. (3) It is possible, but optional, to select rotatable bonds that move the fewest possible atoms, or the largest possible number. If you wish to do this, Select Ligand -> Torsion Tree -> Set Number of Torsions (Figure 7). Figure 7. Setting Number of Torsions in Ligand
You can see how this works by entering different numbers in the data window and alternately clicking the fewest/most buttons while watching the molecule in the display window. When finished manipulating the torsions, click Done. The display now looks like this (Figure 8): Figure 8. Ligand with Torsions Selected
(4) To save the modified file, select Ligand -> Output -> Write PDBQT. Enter a the name using the extension .pdbqt. (For the tutorial, this will be ind.pdbqt.) The Q signifies that charges have been added, and the T that torsions have been selected. Click Save. Then hide the root marker and ligand: click on its gray Show/Hide marker.
V. Flexible Residues in the Protein Beginning with AutoDock 4, support was added for making a portion of the protein flexible, rather than treating it entirely as a rigid shape. A. Redisplay hsg1 by clicking its gray Show/Hide marker. Select Flexible Residues -> Input -> Choose Macromolecule. Then click on sg1 in the widget that appears and click on Select Molecule. You will get a message that Gasteiger charges and AutoDock atom types have been added. (1) If you are asked whether you want to merge the non-polar hydrogens, click OK. B. Select the residues to be flexible: Select -> Select from String. (1) Click Clear Form to remove the waters. Working on the tutorial, type ARG8 in the Residue box, Click Add, and then Dismiss. (2) At the bottom of the Dashboard, the Selected box should show 2 Residues (3) Now define the rotatable bonds in the selected residues: Flexible Residues > Choose Torsions in Currently Selected Residues. Everything except the selected residues disappears, and the Torsion Count widget appears (Figure 9): Figure 9. Torsion Count Widget for Flexible Residues in Protein
In the display, click on the bond between CA and CB in each residue to inactivate it. (You may have to rotate the display in order to see both residues.) Six bonds are now rotatable. Click Close. Clear the Selected box by clicking on the red dots.
(4) Save the flexible residues: Flexible Residues -> Output -> Save Flexible PDBQT. Type hsg1_flex.pdbqt in the File Browser window and Slick Save. (5) Repeat, but save the Rigid PDBQT as hsg1_rigid.pdbqt. C. Remove the original version of hsg1: Edit -> Delete -> Delete Molecule. Choose hsg1, click on Delete Molecule, and click Dismiss. VI. Further Preparation of the Protein Files The next step is to prepare the grid files. Load the first protein file: Grid ->. Macromolecule -> Open. Locate and click on hsg1_rigid.pdbqt. A. ADT checks the charges. If it asks if you want to retain the existing charges, click Yes. If you get a warning about non-integer charges, click OK. Dismiss any other messages that may appear. B. If your ligand file is still open, select the ligand: Grid -> Set Map Types -> Choose Ligand. Select ind. If it is not open use: Grid -> Set Map types -> Open Ligand, and choose ind.pdbqt. Both your protein and your ligand are now displayed. NOTE: The official tutorial at this point suggests that the AutoGpf Ligand widget will open when you select the ligand. It does not. Before going on, use Grid -> Set Map Types -> Directly to check that all atom types in your ligand are represented. If not add the ones that are missing and close the widget. (1) Open the Grid Options widget: Grid -> Grid Box.. It looks like this (Figure 10) You will also notice that a blue and red box has appeared on the main display. You can use the View menu on the Grid Options widget to control the display of the blue box; for example, changing it to display as lines. If you rotate the display, you can see that the box is 3-dimensional (Figure 11). This is the box within which we are going to search; the Grid Box widget allows you to set the size of the box, its location, and the number and spacing of the points within it that will be tested. (2) Use the thumbwheels to set the numbers of points to 60, 60, and 66. You can do this by clicking and dragging on the wheel, or by right-clicking and entering the number in the box that appears. (3) This will center the grid box on the active site of the HIV-1 protease. These numbers, of course, will vary from enzyme to enzyme. They are derived by inspecting the pdb file and locating the coordinates of the active site residue(s).
(4) Use the File menu on the box to Close Saving Current. Then write the grid point file: Grid -> Output -> Save GPF. Name the file hsg1.gpf. C. You are now ready to run AutoGrid. (1) Autogrid can be run from within AutoDock Tools. Choose Run -> AutoGrid from the tan menu. The widget (Figure 12) appears. Figure 10. The Grid Options Widget
Use the Browse button to navigate to /usr/local/AutoDock/autogrid4. Parameter filename should be displayed automatically; Browse to it if it is not. The log filename will be generated from the parameter filename, and the run command in the last box will be displayed.
(2) Run the program by pressing Launch. A small process manager widget will appear that will show you that AutoGrid is running, and allow you to terminate it if desired. AutoGrid will generate a series of Grid Parameter Map files, as seen in the directory listing below (Figure 13).
Figure 11. The Grid Box
D. Next, we prepare the Docking Parameter file.
(1) From the tan menu, choose Docking -> Macromolecule -> Set Rigid Filename, and select hsg1_rigid.pdbqt. (2) Next, choose Docking -> Ligand -> Choose, and select ind A panel appears (below) that allows you to set the initial position of the ligand and initial values for the dihedrals. For the tutorial, simply click Accept. (3) Set the Flexible Residues file: Docking -> Macromolecule -> Set Flexible Residues Filename, and choose hsg1_flex.pdbqt. click Open. Figure 12. The Run AutoGrid Widget
(3) To set the search parameters, choose Docking -> Search Parameters -> Genetic Algorithm. Another widget appears (below). Select the Short setting, which gives 250,000 energy evaluations. Click Accept. Figure 13. Directory Listing Showing Maps Created by AutoGrid.
Figure 14. The dpf File Widget
(4) Choose the random number generator. Docking -> Docking Parameters. Just go with the defaults in this tutorial. (Figure 15)
(5) Set the output filename and the choice of genetic algorithm. Docking -> Output -> Lamarckian GA. Type in ind.dlg and click save.
E. Running AutoDock AutoDock must be run in the directory where the macromolecule, ligand, gpf, dpf, and map files are located. In principle, AutoDock can be started from the Run menu in ADT just the way you started AutoGrid. When I did this, AutoDock ran long enough to write the starting conditions into the log file, and quit. So then I opened a shell window, moving into the directory with the necessary files, and typed at the command line: autodock p ind.dpf l ind.glg & And the program ran properly. When it finished, it printed the message: /usr/local/Autodock/autodock4 Successful Completion on karplus If you used an alias other than autodock for the AutoDock executable, replace autodock in the above command with whatever alias you used.
Figure 15. The Genetic Algorithm Widget
Figure 16. Choosing the Random Number Generator.
Das könnte Ihnen auch gefallen
- Tutorial Docking With GlideDokument5 SeitenTutorial Docking With GlideJohn M. E. PurbaNoch keine Bewertungen
- Essential Algorithms: A Practical Approach to Computer AlgorithmsVon EverandEssential Algorithms: A Practical Approach to Computer AlgorithmsBewertung: 4.5 von 5 Sternen4.5/5 (2)
- Franc3D V6 Ansys Fretting TutorialDokument15 SeitenFranc3D V6 Ansys Fretting Tutorialronald_edinsonNoch keine Bewertungen
- Molecular Modeling of Proteins PDFDokument474 SeitenMolecular Modeling of Proteins PDFFlorAyquiCueva100% (1)
- Dot Point IB Biology - AHL - Kerri Humphreys - Science 2010 PDFDokument50 SeitenDot Point IB Biology - AHL - Kerri Humphreys - Science 2010 PDFJulio Cèsar Torres Hernández100% (1)
- 7 Analysis of A Composite Aircraft Structure Using PCOMPGDokument9 Seiten7 Analysis of A Composite Aircraft Structure Using PCOMPGVinoth BalasubramaniyanNoch keine Bewertungen
- Synthetic Protein Switches Methods and Protocols (Methods in Molecular Biology)Dokument332 SeitenSynthetic Protein Switches Methods and Protocols (Methods in Molecular Biology)phyaravi100% (1)
- Nuclear Magnetic Resonance Spectros PDFDokument310 SeitenNuclear Magnetic Resonance Spectros PDFValemtinoNoch keine Bewertungen
- Gromacs Tutorial For Drug Enzyme ComplexDokument21 SeitenGromacs Tutorial For Drug Enzyme ComplexManoj Kumar RoutNoch keine Bewertungen
- Molecular Docking Using AutoDock and MGL Tools - GirinathDokument23 SeitenMolecular Docking Using AutoDock and MGL Tools - GirinathGirinath Pillai60% (5)
- Phast - Adding A Component From DIPPR PDFDokument22 SeitenPhast - Adding A Component From DIPPR PDFelvithaf100% (1)
- Aspen IQ ModelDokument71 SeitenAspen IQ Modelhaseeb100% (1)
- DOCK6.1 TutorialDokument15 SeitenDOCK6.1 Tutorial@lsreshtyNoch keine Bewertungen
- 105 Machine Learning PaperDokument6 Seiten105 Machine Learning PaperAabda AhmedNoch keine Bewertungen
- 3 Protein Structure-S PDFDokument10 Seiten3 Protein Structure-S PDFNathan Pittman0% (1)
- Molecular Biology of The Cell, 5th EditionDokument82 SeitenMolecular Biology of The Cell, 5th EditionBee Nunes24% (66)
- Asp .Net MVCDokument114 SeitenAsp .Net MVCFernando Torrez100% (3)
- Glucose OxidaseDokument42 SeitenGlucose OxidaseShauqah Mawaddah0% (1)
- Application of Data Mining in BioinformaticsDokument5 SeitenApplication of Data Mining in BioinformaticsYuni ListianaNoch keine Bewertungen
- Fundamentals of Enzymology The Cell and Molecular Biology of Catalytic ProteinsDokument0 SeitenFundamentals of Enzymology The Cell and Molecular Biology of Catalytic Proteinsmonica_elizabeth_35Noch keine Bewertungen
- DockingDokument6 SeitenDockingFrancimauro MoraisNoch keine Bewertungen
- Auto Dock TutorialDokument2 SeitenAuto Dock TutorialParijat DasNoch keine Bewertungen
- Re Cross Docking Tutorial 27aug12Dokument5 SeitenRe Cross Docking Tutorial 27aug12dulNoch keine Bewertungen
- Autodock Tutorial Paso A PasoDokument32 SeitenAutodock Tutorial Paso A PasoFabio CastellanosNoch keine Bewertungen
- ) AutoLigand TutorialDokument26 Seiten) AutoLigand TutorialClaudio Alejandro PereiraNoch keine Bewertungen
- Argus Molecular Docking 1Dokument5 SeitenArgus Molecular Docking 1escherichioNoch keine Bewertungen
- AutoDock TutorialDokument3 SeitenAutoDock Tutorialapi-382392950% (2)
- Protein structure modification using SPDBVDokument10 SeitenProtein structure modification using SPDBVPrayukta PadelkarNoch keine Bewertungen
- 11 Molecular DockingDokument6 Seiten11 Molecular DockingCarissa IslaNoch keine Bewertungen
- Basic Tutorial DockThor 1.0 6Dokument17 SeitenBasic Tutorial DockThor 1.0 6Emmanuel MarinhoNoch keine Bewertungen
- Modul DockingDokument14 SeitenModul DockingSagung DyahNoch keine Bewertungen
- Using Autodock With Autodocktools: A Tutorial: Written by Ruth Huey and Garrett M. MorrisDokument69 SeitenUsing Autodock With Autodocktools: A Tutorial: Written by Ruth Huey and Garrett M. MorrisIsa AlojNoch keine Bewertungen
- Manual BioblenderDokument28 SeitenManual BioblenderFernando Henrique DeslockNoch keine Bewertungen
- Autodock - Vina ProtocolDokument12 SeitenAutodock - Vina ProtocolNaresh kumarNoch keine Bewertungen
- Molecular Docking Tostudy Protein-Ligand InteractionDokument5 SeitenMolecular Docking Tostudy Protein-Ligand InteractionDr. Kaushal Kishor SharmaNoch keine Bewertungen
- 2012 DOCK Tutorial With StreptavidinDokument18 Seiten2012 DOCK Tutorial With Streptavidinboy20_bh100% (1)
- Protein Structure ExplorationDokument22 SeitenProtein Structure Explorationsuveer698Noch keine Bewertungen
- Guidelines and QuestionsDokument6 SeitenGuidelines and Questionsd nNoch keine Bewertungen
- Petrolog ManualDokument23 SeitenPetrolog ManualNibin TomNoch keine Bewertungen
- Institutional Summer Training WorksheetDokument9 SeitenInstitutional Summer Training WorksheetApoorva SinghNoch keine Bewertungen
- Docking TutorialDokument48 SeitenDocking Tutorialapi-3823929100% (4)
- Altera Max Plus TutorialDokument15 SeitenAltera Max Plus TutorialMarwan AhmedNoch keine Bewertungen
- Illustrated Step-By-Step Protocol To Perform Molecular DockingDokument64 SeitenIllustrated Step-By-Step Protocol To Perform Molecular DockingMasyitah ulfayuraNoch keine Bewertungen
- 5700 MlogitDokument38 Seiten5700 MlogitNawaz AliNoch keine Bewertungen
- PyMol PracticalDokument7 SeitenPyMol PracticalTom FlemingNoch keine Bewertungen
- Chimera ProceduresDokument3 SeitenChimera Proceduresraj_shiv10Noch keine Bewertungen
- OS-T - 2040 Spot Weld PDFDokument4 SeitenOS-T - 2040 Spot Weld PDFHrishikesh PhadkeNoch keine Bewertungen
- Modal Transient Dynamic Analysis of A BracketDokument8 SeitenModal Transient Dynamic Analysis of A BracketSrikanth Kabbal MNoch keine Bewertungen
- Arguslab NavodilaDokument6 SeitenArguslab NavodilaChandrakanthNoch keine Bewertungen
- AutoDock Tutorial v1.2Dokument3 SeitenAutoDock Tutorial v1.2Judith NelsonNoch keine Bewertungen
- Using Modal Analysis: ExerciseDokument5 SeitenUsing Modal Analysis: ExercisePraveen SreedharanNoch keine Bewertungen
- Replay A Log On Petri Net For Conformance Analysis Plug-In PDFDokument11 SeitenReplay A Log On Petri Net For Conformance Analysis Plug-In PDFNovandi BanitamaNoch keine Bewertungen
- DSP LAB - IntroductionDokument29 SeitenDSP LAB - IntroductionrkNoch keine Bewertungen
- Dock and Score Tyrosine Kinase InhibitorsDokument14 SeitenDock and Score Tyrosine Kinase InhibitorsvarinderkumarscribdNoch keine Bewertungen
- PyRx Manual PDFDokument13 SeitenPyRx Manual PDFநமச்சிவாயம் பாலகிருஷ்ணன்100% (2)
- Autodock ProtocolDokument17 SeitenAutodock ProtocolSasikala RajendranNoch keine Bewertungen
- Fatigue Analysis (Damage Calculation) Using S-N ApproachDokument7 SeitenFatigue Analysis (Damage Calculation) Using S-N ApproachdhanrajNoch keine Bewertungen
- ADT Autodock TutorialDokument51 SeitenADT Autodock TutorialelmozzNoch keine Bewertungen
- 01 - Lab - 1- Mô phỏng ADSDokument19 Seiten01 - Lab - 1- Mô phỏng ADSNguyễn Ngọc TháiNoch keine Bewertungen
- Input/Edit Core DipsDokument12 SeitenInput/Edit Core DipsKopet GaringNoch keine Bewertungen
- Introduction To Codewarrior™ - Simulating The Microcontroller in Assembly LanguageDokument9 SeitenIntroduction To Codewarrior™ - Simulating The Microcontroller in Assembly LanguageChristian CruzNoch keine Bewertungen
- ChemSep Tutorial: Understanding PCDmanagerDokument37 SeitenChemSep Tutorial: Understanding PCDmanagerjoseNoch keine Bewertungen
- WinHYDROPRO 10.54 user guideDokument4 SeitenWinHYDROPRO 10.54 user guideferdinanfirdausNoch keine Bewertungen
- Getting Started With Postgis: TopicDokument30 SeitenGetting Started With Postgis: TopicSAMINA ATTARINoch keine Bewertungen
- PyrxDokument13 SeitenPyrxahsaanahmadNoch keine Bewertungen
- C++ RefcardDokument2 SeitenC++ RefcardmaxNoch keine Bewertungen
- C++ Short NoteDokument6 SeitenC++ Short Notejohn07bbbNoch keine Bewertungen
- Programming in C Sharp Course Great GuidDokument3 SeitenProgramming in C Sharp Course Great GuidrednriNoch keine Bewertungen
- Power ShellDokument3 SeitenPower ShellrednriNoch keine Bewertungen
- CNF Unit-IV Notes CSETUBE PDFDokument45 SeitenCNF Unit-IV Notes CSETUBE PDFrednriNoch keine Bewertungen
- Blg252e CompleteDokument561 SeitenBlg252e Completesuleman247Noch keine Bewertungen
- C++ Neural Networks and Fuzzy Logic - Valluru B. RaoDokument595 SeitenC++ Neural Networks and Fuzzy Logic - Valluru B. RaoThyago VasconcelosNoch keine Bewertungen
- C++ Notes on OOPDokument133 SeitenC++ Notes on OOPrednri0% (1)
- C Language Standard Data Types and OperatorsDokument2 SeitenC Language Standard Data Types and OperatorsPradeep KumarNoch keine Bewertungen
- Html5 TutorialDokument23 SeitenHtml5 TutorialHands OffNoch keine Bewertungen
- Cil2012 html5Dokument45 SeitenCil2012 html5sreejeshkeralamNoch keine Bewertungen
- CNF Unit-I Notes Csetube PDFDokument37 SeitenCNF Unit-I Notes Csetube PDFthebhas1954Noch keine Bewertungen
- What You'll Build: by Scott Hanselman - January 12, 2011Dokument334 SeitenWhat You'll Build: by Scott Hanselman - January 12, 2011Ebenezer Tweneboah-KoduahNoch keine Bewertungen
- Adafruit Pitft 3 Dot 5 Touch Screen For Raspberry Pi PDFDokument50 SeitenAdafruit Pitft 3 Dot 5 Touch Screen For Raspberry Pi PDFrednri100% (1)
- 2609a Delivery Guide PDFDokument748 Seiten2609a Delivery Guide PDFrednriNoch keine Bewertungen
- AJAX Weather, JQuery Effects, AngularJS City AppDokument4 SeitenAJAX Weather, JQuery Effects, AngularJS City ApprednriNoch keine Bewertungen
- ExercisesDokument3 SeitenExercisesrednriNoch keine Bewertungen
- 15 Introduction of Softcomputing Approach in Slope StabilityDokument8 Seiten15 Introduction of Softcomputing Approach in Slope StabilityrednriNoch keine Bewertungen
- 09 - Chapter 3 PDFDokument80 Seiten09 - Chapter 3 PDFrednriNoch keine Bewertungen
- ASP NET 4 and Visual Studio 2010 Web Development OverviewDokument66 SeitenASP NET 4 and Visual Studio 2010 Web Development OverviewKalai VananNoch keine Bewertungen
- ExercisesDokument3 SeitenExercisesrednriNoch keine Bewertungen
- Developing Algorithmic and Logical Thinking SkillsDokument11 SeitenDeveloping Algorithmic and Logical Thinking SkillsrednriNoch keine Bewertungen
- The STRCHR Function: Using STRCHR To Search A String For A Single CharacterDokument20 SeitenThe STRCHR Function: Using STRCHR To Search A String For A Single CharacterrednriNoch keine Bewertungen
- Hidden Markov Models in Bioinformatics: Example Domain: Gene FindingDokument32 SeitenHidden Markov Models in Bioinformatics: Example Domain: Gene FindingrednriNoch keine Bewertungen
- Introduction To S AsDokument16 SeitenIntroduction To S AsrednriNoch keine Bewertungen
- SAS Guide Revf10Dokument12 SeitenSAS Guide Revf10rednriNoch keine Bewertungen
- A Brief Introduction To SAS Operators and FunctionsDokument7 SeitenA Brief Introduction To SAS Operators and FunctionsrednriNoch keine Bewertungen
- Biomolecules and Its UsesDokument20 SeitenBiomolecules and Its UsesRajendra Swarnakar25% (4)
- In Silico Study of Single Chain Fragment Variable (SCFV) On Chikungunya Virus Using Indonesian StrainDokument5 SeitenIn Silico Study of Single Chain Fragment Variable (SCFV) On Chikungunya Virus Using Indonesian StrainAdriani HasyimNoch keine Bewertungen
- Full Download Test Bank For Molecular Cell Biology Eighth Edition PDF Full ChapterDokument36 SeitenFull Download Test Bank For Molecular Cell Biology Eighth Edition PDF Full Chapterscottthorntongjpnfeatrw100% (18)
- A Simple Method For Displaying The Hydropathic Character of A ProteinDokument28 SeitenA Simple Method For Displaying The Hydropathic Character of A ProteinBiosynthesisNoch keine Bewertungen
- Proteins: Their Biological Functions and Primary StructureDokument59 SeitenProteins: Their Biological Functions and Primary StructureIsha BhartiNoch keine Bewertungen
- Structural Changes in Cellulose During PDokument8 SeitenStructural Changes in Cellulose During ParjunanpnNoch keine Bewertungen
- Biology HL Past Papers GuideDokument90 SeitenBiology HL Past Papers GuideelenaNoch keine Bewertungen
- Biochemistry Check for Understanding QuestionsDokument6 SeitenBiochemistry Check for Understanding QuestionsG INoch keine Bewertungen
- CV Alberto PerezDokument5 SeitenCV Alberto Perezaenon99Noch keine Bewertungen
- Chapter 4 Protein Three-Dimensional StructureDokument24 SeitenChapter 4 Protein Three-Dimensional StructureRO OMNoch keine Bewertungen
- 2001 Dobson Gerrard Pratt Foundations of Chemical BiologyDokument110 Seiten2001 Dobson Gerrard Pratt Foundations of Chemical BiologyElmer L'éléphantNoch keine Bewertungen
- Vanders Human Physiology The Mechanisms of Body Function 13th Edition Widmaier Solutions ManualDokument26 SeitenVanders Human Physiology The Mechanisms of Body Function 13th Edition Widmaier Solutions ManualJadeFischerqtcj100% (29)
- UniTartuCS Poster Template PortraitDokument1 SeiteUniTartuCS Poster Template PortraitManuel ZumbadoNoch keine Bewertungen
- Lenoir FlowProcessFold PDFDokument29 SeitenLenoir FlowProcessFold PDFMohammed Gism AllahNoch keine Bewertungen
- A Review On New Target Insulysine (Ide) & Its Inhibitors As Antidiabetics AgentsDokument12 SeitenA Review On New Target Insulysine (Ide) & Its Inhibitors As Antidiabetics AgentsTJPRC PublicationsNoch keine Bewertungen
- Halder2020 hydrophilicAminoAcidsProteinEthanolDokument10 SeitenHalder2020 hydrophilicAminoAcidsProteinEthanolSBNoch keine Bewertungen
- Accurate Calculation of Free Energy Changes Upon Amino Acid MutationDokument29 SeitenAccurate Calculation of Free Energy Changes Upon Amino Acid MutationThales FreireNoch keine Bewertungen
- CHEMICALS OF LIFE @SCZ Salongo 2016 PDFDokument18 SeitenCHEMICALS OF LIFE @SCZ Salongo 2016 PDFKlint Rodney Mark LuleNoch keine Bewertungen
- Proteins: Ms. Jirehkriza G. Suganob General Biology 1Dokument41 SeitenProteins: Ms. Jirehkriza G. Suganob General Biology 1Jcob SntosNoch keine Bewertungen
- Biotechnology SyllabusDokument91 SeitenBiotechnology Syllabuskiran.rNoch keine Bewertungen
- Protein StructureDokument11 SeitenProtein Structurelol heyNoch keine Bewertungen
- Aquasome: A Novel Drug DeliveryDokument11 SeitenAquasome: A Novel Drug DeliveryIjupbs IjupbsNoch keine Bewertungen