Beruflich Dokumente
Kultur Dokumente
T Coffee Technical
Hochgeladen von
JulianGiraldoOriginalbeschreibung:
Originaltitel
Copyright
Verfügbare Formate
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenCopyright:
Verfügbare Formate
T Coffee Technical
Hochgeladen von
JulianGiraldoCopyright:
Verfügbare Formate
CENTRE NATIONAL DE LA RECHERCHE
SCIENTIFIQUE (
T-Coffee code can be re-used freely
Our philosophy is that code is meant to be re-used, including ours. No permission is
needed for the cut and paste of a few functions, although we are always happy to
receive pieces of improved code.
T-Coffee can be incorporated in most pipelines:
Plug-in/Plug-out
Our philosophy is to insure that as many methods as possible can be used as plug-
ins within T-Coffee. Likewise, we will give as much support as possible to anyone
wishing to turn T-Coffee into a plug-in for another method. or more details on how
to do this, see the plug-in and the plug-out sections of the Tutorial !anual.
"gain, you do not need our permission to either use T-Coffee #or your method as a
plug-in$out% but if you let us know, we will insure the stability of T-Coffee within
your system through future releases.
The current license only allows for the incorporation of T-Coffee in non-commercial
pipelines #i.e. where you do not sell the pipeline, or access to it%. &f your pipeline is
commercial, please get in touch with us.
Technical
1
Addresses and
Contacts
Contributors
T-cofee is developed, mai!aied, moi!o"ed, #sed ad de$#%%ed $& a
dedica!ed !eam !'a! icl#de o" 'ave icl#ded(
C)d"ic No!"edame, Fa$"ice A"mo#%om, Des Hi%%is, Se$as!ie
*o"e!!i, O"la O+S#lliva, Eamo O+Toole, Olivie" -oi"o!, .a"s!e
S#'"e, Iai /allace, Ad"eas /ilm
Addresses
'e are always very eager to get some user feedback. (lease do not hesitate to drop
us a line at) cedric.notredame*europe.com The latest updates of T-Coffee are
always available on) www.tcoffee.org . On this address you will also find a link to
some of the online T-Coffee servers, including Tcoffee*igs
T-Coffee can be used to automatically check if an updated version is available,
however the program will not update automatically, as this can cause endless
reproducibility problems.
PROMPT: t_coffee update
2
Citations
&t is important that you cite T-Coffee when you use it. Citing us is #almost% like
giving us money) it helps us convincing our institutions that what we do is useful
and that they should keep paying our salaries and deliver +onuts to our offices from
time to time #Not that they ever did it, but it would be nice anyway%.
Cite the server if you used it, otherwise, cite the original paper from ,--- #No, it
was never named .T-Coffee ,---.%.
Notredame C, /iggins +0,
/eringa 1.
2elated "rticles, Links
T-Coffee) " novel method for fast and accurate multiple se3uence
alignment.
1 !ol 4iol. ,--- 5ep 678-,#9%),-:-9;.
(!&+) 9-<=>:;- ?(ub!ed - inde@ed for !A+L&NAB
Other useful publications include)
T-Coffee
Claude 14, 5uhre C,
Notredame C, Claverie 1!,
"bergel C.
2elated "rticles, Links
Casp2) a web server for automated molecular replacement using
homology modelling.
Nucleic "cids 2es. ,--> 1ul 978,#'eb 5erver issue%)'=-=-<.
(!&+) 9:,9:>=- ?(ub!ed - inde@ed for !A+L&NAB
(oirot O, 5uhre C, "bergel C,
ODToole A, Notredame C.
2elated "rticles, Links
8+Coffee*igs) a web server for combining se3uences and
structures into a multiple se3uence alignment.
Nucleic "cids 2es. ,--> 1ul 978,#'eb 5erver issue%)'8;->-.
(!&+) 9:,9:8>: ?(ub!ed - inde@ed for !A+L&NAB
OD5ullivan O, 5uhre C,
2elated "rticles, Links
3
"bergel C, /iggins +0,
Notredame C.
8+Coffee) combining protein se3uences and structures within
multiple se3uence alignments.
1 !ol 4iol. ,--> 1ul ,78>-#,%)86:-<:.
(!&+) 9:,-9-:< ?(ub!ed - inde@ed for !A+L&NAB
(oirot O, ODToole A,
Notredame C.
2elated "rticles, Links
Tcoffee*igs) " web server for computing, evaluating and
combining multiple se3uence alignments.
Nucleic "cids 2es. ,--8 1ul 9789#98%)8:-8-=.
(!&+) 9,6,>8:> ?(ub!ed - inde@ed for !A+L&NAB
Notredame C. 2elated "rticles, Links
!occa) semi-automatic method for domain hunting.
4ioinformatics. ,--9 "pr79;#>%)8;8->.
(!&+) 998-98-< ?(ub!ed - inde@ed for !A+L&NAB
Notredame C, /iggins +0,
/eringa 1.
2elated "rticles, Links
T-Coffee) " novel method for fast and accurate multiple se3uence
alignment.
1 !ol 4iol. ,--- 5ep 678-,#9%),-:-9;.
(!&+) 9-<=>:;- ?(ub!ed - inde@ed for !A+L&NAB
Notredame C, /olm L,
/iggins +0.
2elated "rticles, Links
COAA) an obEective function for multiple se3uence alignments.
4ioinformatics. 9<<6 1un79>#:%)>-;-,,.
(!&+) <=6,-:> ?(ub!ed - inde@ed for !A+L&NAB
occa
Notredame C. 2elated "rticles, Links
!occa) semi-automatic method for domain hunting.
4ioinformatics. ,--9 "pr79;#>%)8;8->.
(!&+) 998-98-< ?(ub!ed - inde@ed for !A+L&NAB
C!"#
http)$$www.tcoffee.org$(ublications$(df$core.pp.pdf
$
!ther Contributions
'e do not mean to steal code, but we will always try to re-use pre-e@isting code
whenever that code e@ists, free of copyright, Eust like we e@pect people to do with
our code. /owever, whenever this happens, we make a point at properly citing the
source of the original contribution. &f ever you recogniFe a piece of your code
improperly cited, please drop us a note and we will be happy to correct that.
&n the mean time, here are some important pieces of code from other packages that
have been incorporated within the T-Coffee package. These include)
-The 5im algorithm of /uang and !iller that given two se3uences computes
the N best scoring local alignments.
-The tree reading$computing routines are taken from the Clustal' (ackage,
courtesy of 1ulie Thompson, +es /iggins and Toby 0ibson #Thompson, /iggins,
0ibson, 9<<>, >=;8->=6-,vol. ,,, Nucleic "cid 2esearch%.
-The implementation of the algorithm for aligning two se3uences in linear
space was adapted from !yers and !iller, in C"4&O5, 9<66, 99-9;, vol. 9%
-Garious techni3ues and algorithms have been implemented. 'henever
relevant, the source of the code$algorithm$idea is indicated in the corresponding
function.
-=> 4its compliance was implemented by 4enEamin 5ohn, (erformance
Computing Center 5tuttgart #/L25%, 0ermany
-+avid !athog #Caltech% provided many fi@es and useful feedback for
improving the code and making the whole soft behaving more rationally
%ug "eports and &eedbac'
-(rof +avid 1ones #HCL% reported and corrected the (+49C bug #now
tIcoffee$sap can align (+4 se3uences longer than 9--- ""%.
-1ohan Leckner reported several bugs related to the treatment of (+4
structures, insuring a consistent behavior between version 9.8; and current ones.
(
)nstallation of The T-
Coffee Pac'ages
Third Party Pac'ages and !n *emand
)nstallations
T-Coffee is a comple@ package that interacts with many other third part software. &f
you only want a standalone version of T-Coffee, you may install that package on its
own. &f you want to use a most sophisticated flavor #8dcoffee, e@presso, rcofeee,
etc...%, the installer will try to install all the third party packages re3uired.
Note that since version ;.:=, T-Coffee will use Don demandD installation and install
the third party packages it needs JwhenJ it needs them. This only works for
packages not re3uiring specific licenses and that can be installed by the regular
installer. (lease let us know if you would like another third party package to be
included.
'henver on-demand installation or automated installation fails because of
unforessen system specificities, users should install the third party package
manually. This documentation gives some tips we have found useful, but users are
encouraged to check the original documentation.
+tandard )nstallation of T-Coffee
,ni-
Kou need to have) gcc, g;;, C("N and an internet connection and your root
password #to install 5O"(%. &f you cannot log as root, ask #kindly% your system
manager to install 5O"())Lite for you. Kou may do this before or after the
installation of T-Coffee. Aven without 5O"( you will still be able to use the basic
functions of T-Coffee #simplest usage%.
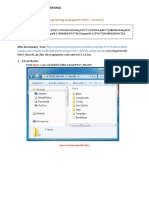
0, gunzip t_coffee.tar.gz
1, tar -xvf t_coffee.tar
2, cd t_coffee
3, ./install t_coffee
This installation will only install the stand alone T-Coffee. &f you want to install a
.
specific mode of T-Coffee, you may try the following commands that will try to
gather all the necessary third party packages. Note that a package already found on
your system will not be re-installed.
./install t_coffee
./install mcoffee
./install 3dcoffee
./install rcoffee
./install psicoffee
Or even
./install all
-"ll the corresponding e@ecutables will be downloaded automatically and installed
in
$HOME/.t_coffee/plugins
-if you e@ecutables are in a different location, give it to T-Coffee using the -plugins
flag.
-&f the installation of any of the companion package fails, you should install it
yourself using the provided link #see below% and following the authors instructions.
-&f you have not managed to install 5O"())Lite, you can re-install it later #from
anywhere% following steps 9-,.
-This procedure attempts 8 things) installing and Compiling T-Coffee #C program%,
&nstalling and compiling T!align #ortran%, &nstalling and compiling
5O"())Lite#(erl !odule%.
-&f you have never installed 5O"())Lite, C("N will ask you many 3uestions) say
Kes to all
-&f everything went well, the procedure has created in the bin directory two
e@ecutables) tIcoffee and T!align #Make sure these executables are on your
$PATH!%
/
icrosoft 0indo1s/Cyg1in
&nstall Cygwin
+ownload The &nstaller #NOT Cygwin$L%
Click on view to list "LL the packages
5elect) gcc-core, make, wget
Optional) ssh, @emacs, nano
2un mkpasswd in Cywin #as re3uested when you start cygwin%
&nstall T-Coffee within Cygwin using the Hni@ procedure
AC os23 4inu-
!ake sure you have the +eveloperDs kit installed #compilers and makefile%
ollow the Hni@ (rocedure
C4,+T#" )nstallation
&n order to run, T-Coffee must have a value for the httpIpro@y and for the A-mail. &n
order to do so you can either)
e@port the following values)
e@port httpIpro@yI>ITCOAAM.pro@y. or .. if no pro@y
e@port A!"&LI>ITCOAAM.your email.
O2
modify the file N$.tIcoffee$tIcoffeeIenv
O2
add to your command line) tIcoffee O. -pro@yMPpro@yQ -emailMPemail
if you have no pro@y) tIcoffee O -pro@y -emailMPemailQ
)f you ha5e P*% installed:
"ssuming you have a standard (+4 installation in your file system
seten !or e"port# P$%_$&R 'a(s
pat)*/data/structures/all/pd(/
OR
seten !or e"port# P$%_$&R 'a(s
pat)*/structures/diided/pd(/
&f you do not have (+4 installed, donDt worry, tIcoffee will go and fetch any
structure it needs directly from the (+4 repository. &t will simply be a bit slower
than if you had (+4 locally.
6
)nstalling %4A+T for T-Coffee
4L"5T is a program that search se3uence databases for homologues of a 3uery
se3uence. &t works for proteins and Nucleic "cids. &n theory 4L"5T is Eust a
package like any, but in practice things are a bit more comple@. To run well, 4L5T
re3uires up to date databases #that can be fairly large, like N2 or HN&(2OT% and a
powerful computer.
ortunately, an increasing number of institutes or companies are now providing
4L"5T clients that run over the net. &t means that all you need is a small program
that send your 3uery to the big server and gets the results back. This prevents you
from the hassle of installing and maintaining 4L"5T, but of course it is less private
and you rely on the network and the current load of these busy servers.
Thanks to its interaction with 4L"5T, T-Coffee can gather structures and protein
profiles and deliver an alignment significantly more accurate than the default you
would get with T-Coffee or any similar method.
Let us go through the various modes available for T-Coffee
0hy *o ) need %4A+T 1ith T-Coffee7
The most accurate modes of T-Coffe scan the databases for templates that they use
to align the se3uences. There are currently two types of templates for proteins)
structures #(+4% that can be found by a blastp against the (+4 database and
profiles that can be constructed with eiether a blastp or a psiblast against nr or
uniprot.
These templates are automatically built if you use)
t_coffee '+ourse,* -mode e"presso
that fetches aand uses pdb templates, or
t_coffee '+our se,* -mode psicoffee
that fetches and uses profile templates, or
t_coffee '+our se,* -mode accurate
that does everything and tries to use the best template. Now that you see why
it is useful letDs see how to get 4L"5T up and running, from the easy solution to
tailor made ones.
,sing the #%) %4A+T Client
This is by far the easiest #and the default mode%. The perl clients are already
incorporated in T-Coffeem and all you need is the 5O"())Lite perl library. &n theory,
T-Coffee should have already installed this library during the standard installation.
8
Ket, this re3uires having toot access. &f you did not have it at the time of the
installation, or if you need your system administrator to install 5O"())Lite, simply
follow the instruction provided on the website)
)ttp://searc).cpan.org/.(+rne//O0P-1ite-2.32a
&t really is worth the effort, since the A4& is providing one of the best webservice
available around, and most notably, the only public psiblast via a web service.
"nother important point is that the A4& re3uires your A-mail address to process your
3ueries. Normally, T-Coffee should have asked you to provide this address. &f you
have not, or if you have provided a phony address, you should correct this by
directly editing the file
./.t_coffee/email.t"t
Be Careful! &f you provide a fake A-mail, the A4& may suspend the service for all
machines associated with your &( address #that could mean your entire lab, or entire
institute, or even the entire country or, but & doubt it, the whole universe%.
,sing the 9C%) %4A+T Client
The NC4& is the ne@t best alternative. &n my hand it was always a bit slower and
most of all, it does not incorporate (5&-4L"5T #as a web sevice%. " big miss. The
NC4& web blast client is a small e@ecutable that you should install on your system
following the instructions given on this link
ftp://ftp.nc(i.ni).go/(last/e"ecuta(les/10TE/T
5imply go for netbl, download the e@ecutable that corresponds to your architecture
#cygwin users should go for the win e@ecutable%. +espite all the files that come
along the e@ecutable blastcl8 is a stand alone e@ecutable that you can safely move to
your R4&N.
"ll you will then need to do is to make sure that T-Coffee uses the right client, when
you run it.
-(last_serer456%&
No need for any A-mail here, but you donDt get psiblast, and whenever T-Coffee
wants to use it, blastp will be used instead.
,sing another Client
Kou may have your own client #lucky you%. &f that is so, all you need is to make sure
that this client is complient with the blast command line. &f your client is named
foo.pl, all you need to to is run T-Coffee with
1:
-(last_serer461&E5T_foo.pl
oo will be called as if it were blastpgp, and it is your responsability to make sure it
can handle the following command line)
foo.pl -p 'met)od* -d 'd(* -i 'infile* -o 'outfile* -m 7
method can either be blastp or psiblast.
infile is a "5T" file
-m; triggers the L!L output. T-Coffee is able to parse both the A4& L!L output
and the NC4& L!L output.
&f foo.pl behaves differently, the easiest will probably be to write a wrapper around
it so that wrappedIfoo.pl behaves like blastpgp
,sing a %4A+T local 5ersion on ,9)2
&f you have blastpgp installed, you can run it instead of the remote clients by using)
-(last_serer41O601
The documnentation for blastpgp can be found on)
888.nc(i.nlm.ni).go/staff/tao/9R10P&/(lastpgp.)tml
and the package is part of the standard 4L"5T distribution
ftp://ftp.nc(i.ni).go/(last/e"ecuta(les/10TE/T
+epending on your system, your own skills, your re3uirements and on more
parameters than & have fingers to count, installing a 4L"5T server suited for your
needs can range from a 9- minutes Eob to an achivement spread over several
generations. 5o at this point, you should roam the NC4& website for suitable
information.
&f you want to have your own 4L"5T server to run your own databases, you should
know that it is possible to control both the database and the program used by
4L"5T)
-protein_db: will specify the database used by all the psi-blast
modes
-pdb_db: will specify the database used by the pdb modes
11
,sing a %4A+T local 5ersion on 0indo1s/cyg1in
or those of you using cygwin, be careful. 'hile cygwin behaves like a HN&L
system, the 4L"5T e@ecutable re3uired for cygwin #win8,% is e@pecting
'&N+O'5 path and not HN&L path. This has three important conse3uences)
9- the ncbi file declaring the +ata directory must be)
C)'&N+O'5$$ncbi.init ?at the root of your '&N+O'5B
,- the address mentionned with this file must be '&N+O'5 formated, for instance,
on my system)
+ataMC)ScygwinShomeSnotredameSblastSdata
8- 'hen you pass database addresses to 4L"5T, these must be in 'indows format)
-proteinIdbM.c)$somewhere$somewhereelse$database.
#using the slash #$% or the andtislash #S% does not matter on new systems but & would
reommand against incorporating white spaces.
)nstalling !ther Companion Pac'ages
T-Coffee is meant to interact with as many packages as possible, either for aligning
or using predictions. &f you type
t_coffee
Kou will receive a list of supported packages that looks like the ne@t table. &n theory,
most of these packages can be installed by T-Coffee
****** Pairwise e!uence "lignment #ethods:
--------------------------------------------
fast_pair built_in
exon$_pair built_in
exon%_pair built_in
exon_pair built_in
slow_pair built_in
proba_pair built_in
lalign_id_pair built_in
se!_pair built_in
externprofile_pair built_in
hh_pair built_in
profile_pair built_in
cdna_fast_pair built_in
cdna_cfast_pair built_in
clustalw_pair ftp://www.ebi.ac.u&/pub/clustalw
mafft_pair http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
mafft'tt_pair http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
mafftgins_pair http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
dialigntx_pair http://dialign-tx.gobics.de/
dialignt_pair http://dialign-t.gobics.de/
poa_pair http://www.bioinformatics.ucla.edu/poa/
probcons_pair http://probcons.stanford.edu/
12
muscle_pair http://www.drive).com/muscle/
t_coffee_pair http://www.tcoffee.org
pcma_pair ftp://iole.swmed.edu/pub/P*#"/
&align_pair http://msa.cgb.&i.se
amap_pair http://bio.math.ber&eley.edu/amap/
proda_pair http://bio.math.ber&eley.edu/proda/
pran&_pair http://www.ebi.ac.u&/goldman-srv/pran&/
consan_pair http://selab.'anelia.org/software/consan/
****** Pairwise tructural "lignment #ethods:
--------------------------------------------
align_pdbpair built_in
lalign_pdbpair built_in
extern_pdbpair built_in
thread_pair built_in
fugue_pair http://www-
cryst.bioc.cam.ac.u&/fugue/download.html
pdb_pair built_in
sap_pair http://www-
cryst.bioc.cam.ac.u&/fugue/download.html
mustang_pair http://www.cs.mu.oz.au/(arun/mustang/
tmalign_pair http://zhang.bioinformatics.&u.edu/+#-align/
****** #ultiple e!uence "lignment #ethods:
--------------------------------------------
clustalw_msa ftp://www.ebi.ac.u&/pub/clustalw
mafft_msa http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
mafft'tt_msa http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
mafftgins_msa http://www.biophys.&yoto-
u.ac.'p/(&atoh/programs/align/mafft/
dialigntx_msa http://dialign-tx.gobics.de/
dialignt_msa http://dialign-t.gobics.de/
poa_msa http://www.bioinformatics.ucla.edu/poa/
probcons_msa http://probcons.stanford.edu/
muscle_msa http://www.drive).com/muscle/
t_coffee_msa http://www.tcoffee.org
pcma_msa ftp://iole.swmed.edu/pub/P*#"/
&align_msa http://msa.cgb.&i.se
amap_msa http://bio.math.ber&eley.edu/amap/
proda_msa http://bio.math.ber&eley.edu/proda/
pran&_msa http://www.ebi.ac.u&/goldman-srv/pran&/
,,,,,,, Prediction #ethods available to generate +emplates
-------------------------------------------------------------
-."plfold http://www.tbi.univie.ac.at/(ivo/-."/
/##top www.enzim.hu/hmmtop/
01-2 http://mig.'ouy.inra.fr/logiciels/gor34/
wublast_client
http://www.ebi.ac.u&/+ools/webservices/services/wublast
blastpgp_client
http://www.ebi.ac.u&/+ools/webservices/services/blastpgp
5555555555555555555555555555555555555555555555555555555555
13
)nstallation of P+)-Coffee and #-presso
(5&-Coffee is a mode of T-Coffee that runs a a (si-4L"5T on each of your
se3uences and makes a multiple profile alignment. &f you do not have any structural
information, it is by far the most accurate mode of T-Coffee. To use it, you must
have 5O"( installed so that the A4& 4L"5T client can run on your system.
&t is a bit slow, but really worth it if your se3uences are hard to align and if the
accuracy of your alignment is important.
To use this mode, try)
t_coffee '+ourse,uence* -mode psicoffee
Note that because (5&-4L"5T is time consuming, T-Coffee stores the runs in its
cache #.$tcoffee$cache% so that it does not need to be re-run. &t means that if you re-
align your se3uences #or add a few e@tra se3uences%, things will be considerably
faster.
&f your installation procedure has managed to compile T!align, and if T-Coffee has
access to the A4& 4L"5T server #or any other server% you can also do the following)
t_coffee '+ourse,uence* -mode e"presso
That will look for structural templates. "nd if both these modes are running fine,
then you are ready for the best, the .crTme de la crTme.)
t_coffee '+ourse,uence* -mode accurate
)nstallation of -Coffee
!-Coffee is a special mode of T-Coffee that makes it possible to combine the output
of many multiple se3uence alignment packages.
Automated )nstallation
&n the T-Coffee distribution, type)
./install mcoffee
&n theory, this command should download and install every re3uired package. &f,
however, it fails, you should switch to the manual installation #see ne@t%.
4y default these packages will be in
$HOME/.t_coffee/plugins
1$
&f you want to have these companion packages in a different directory, you can
either set the environement variable
P19:&5/_;_T6O<<EE4'plugins dir*
Or use the command line flag -plugin #over-rides every other setting%
t_coffee ... -plugins4'plugins dir*
anual )nstallation
!-Coffee re3uires a standard T-Coffee installation #c.f. previous section% and the
following packages to be installed on your system)
Pac&age 6here 7rom
5555555555555555555555555555555555555555555555555555555555
*lustal6 can interact with t_coffee
----------------------------------------------------------
Poa http://www.bioinformatics.ucla.edu/poa/
----------------------------------------------------------
#uscle http://www.drive).com
----------------------------------------------------------
Prob*ons http://probcons.stanford.edu/
Prob*ons-." http://probcons.stanford.edu/
----------------------------------------------------------
#"77+ http://www.biophys.&yoto-u.ac.'p/(&atoh/programs/align/mafft/
----------------------------------------------------------
8ialign-+ http://dialign-t.gobics.de/
8ialign-+9 http://dialign-tx.gobics.de/
----------------------------------------------------------
P*#" ftp://iole.swmed.edu/pub/P*#"/
----------------------------------------------------------
&align http://msa.cgb.&i.se
----------------------------------------------------------
amap http://bio.math.ber&eley.edu/amap/
-----------------------------------------------------------
proda_msa http://bio.math.ber&eley.edu/proda/
-----------------------------------------------------------
pran&_msa http://www.ebi.ac.u&/goldman-srv/pran&/
&n our hands all these packages where very straightforward to compile and install on
a standard cygwin or Linu@ configuration. 1ust make sure you have gcc, the C
compiler, properly installed.
Once the package is compiled and ready to use, make sure that the e@ecutable is on
your path, so that tIcoffee can find it automatically. Our favorite procedure is to
create a bin directory in the home. &f you do so, make sure this bin is in your path
and fill it with all your e@ecutables #this is a standard Hni@ practice%.
1(
&f for some reason, you do not want this directory to be on your path, or you want to
specify a precise directory containing the e@ecutables, you can use)
e"port P19:&5/_;_T6O<<EE4'dir*
4y default this directory is set to R/O!A$.tIcoffee$plugins$RO5, but you can over-
ride it with the environement variable or using the flag)
t_coffee ...-plugins4'dir*
&f you cannot, or do not want to use a single bin directory, you can set the following
environment variables to the absolute path values of the e@ecutable you want to use.
'henever they are set, these variables will supersede any other declaration. This is
a convenient way to e@periment with multiple package versions.
PO0_;_T6OO<<EE
619/T01=_;_T6O<<EE
PO0_;_T6O<<EE
T6O<<EE_;_T6O<<EE
M0<<T_;_T6O<<EE
M9/61E_;_T6O<<EE
$&01&:5T_;_T6O<<EE
PR05>_;_T6O<<EE
$&01&:5T?_;_T6O<<EE
or three of these packages, you will need to copy some of the files in a special T-
Coffee directory.
cp PO0_$&R/@ ./.t_coffee/mcoffee/
cp $&01&:5-T/conf/@ ./.t_coffee/mcoffee
cp $&01&:5-T?/conf/@ ./.t_coffee/mcoffee
Note that the following files are enough for default usage)
%1O/9M.diag_pro(_tA2 %1O/9M7B.scr (losumC2_trunc.mat
dna_diag_pro(_A22_e"p_332222 dna_diag_pro(_D22_e"p_AA2222
%1O/9M.scr %1O/9ME2.scr dna_diag_pro(_A22_e"p_AA2222
dna_diag_pro(_A22_e"p_BB2222 dna_diag_pro(_DB2_e"p_AA2222
%1O/9M7B.diag_pro(_tD (losumC2.mat dna_diag_pro(_A22_e"p_DD2222
1.
dna_diag_pro(_AB2_e"p_AA2222 dna_matri".scr
&f you would rather have the mcoffee directory in some other location, set the
!COAAI>ITCOAA environement variable to the propoer directory)
seten M6O<<EE_;_T6O<<EE 'director+ containing mcoffee files*
)nstallation of AP*% and i"+*
A-D4 ad iR*SD a"e ico"po"a!ed i T-Cofee, Oce !5cofee is
is!alled, &o# ca ivo6#e !'ese p"o%"ams $& !&pi%(
t_coffee ot)er_pg apd(
t_coffee ot)er_pg irmsd
)nstallation of t"+*
!R*SD comes alo% 7i!' !5cofee $#! i! also "e6#i"es !'e pac8a%e
p'&lip i o"de" !o $e 9#c!ioal, -'&lip ca $e o$!aied 9"om(
Pac&age 7unction
555555555555555555555555555555555555555555555555555
---------------------------------------------------
Phylip Phylogenetic tree computation
evolution.genetics.washington.edu/phylip.html
---------------------------------------------------
t_coffee ot)er_pg trmsd
)nstallation of se;<reformat
Se65"e9o"ma! is a "e9o"ma!!i% pac8a%e !'a! is pa"! o9 !5cofee, To #se
i! (ad see !'e availa$le op!ios:, !&pe(
t_coffee ot)er_pg se,_reformat
)nstallation of e-tract<from<pdb
E;!"ac!59"om5pd$ is a -D4 "e9o"ma!!i% pac8a%e !'a! is pa"! o9
!5cofee, To #se i! (ad see !'e availa$le op!ios:, !&pe,
1/
t_coffee ot)er_pg apd( )
E;!"ac!59"om5pd$ "e6#i"es 7%e! i o"de" !o a#!oma!icall& 9e!c' -D4
s!"#c!#"es,
)nstallation of 3*-Coffee/#-presso
8+-Coffee$A@presso is a special mode of T-Coffee that makes it possible to combine
se3uences and structures. The main difference between A@presso and 8+-Coffee is
that A@presso fetches the structures itself.
Automated )nstallation
&n the T-Coffee distribution, type)
./install e"presso
OR
./install 3dcoffee
&n theory, this command should download and install every re3uired package
#except fugue%. &f, however, it fails, you should switch to the manual installation
#see ne@t%.
anual )nstallation
&n order to make the most out of T-Coffee, you will need to install the following
packages #make sure the e@ecutable is named as indicated below%)
Pac&age 7unction
555555555555555555555555555555555555555555555555555
---------------------------------------------------
wget $8*offee
"utomatic 8ownloading of tructures
---------------------------------------------------
sap structure/structure comparisons
:obtain it from 6. +aylor; .3#--#-*<.
---------------------------------------------------
+#align zhang.bioinformatics.&u.edu/+#-align/
---------------------------------------------------
mustang www.cs.mu.oz.au/(arun/mustang/
---------------------------------------------------
wublastclient www.ebi.ac.u&/+ools/webservices/clients/wublast
---------------------------------------------------
=last www.ncbi.nih.nlm.gov
---------------------------------------------------
7ugue* protein to structure alignment program
http://www-cryst.bioc.cam.ac.u&/fugue/download.html
***.1+ *1#P>?1-@***
16
Once the package is installed, make sure make sure that the e@ecutable is on your
path, so that tIcoffee can find it automatically.
The wublast client makes it possible to run 4L"5T at the A4& without having to
install any database locally. &t is an ideal solution if you are only using e@presso
occasionally.
)nstalling &ugue for T-Coffee
Hses a standard fugue installation. Kou only need to install the following packages)
Eoy, melody, fugueali, sstruc, hbond
&f you have root privileges, you can install the common data in)
cp fugue$classdef.dat $data$fugue$5H45T$classdef.dat
otherwise
5etenv !ALO+KICL"55+AMPlocationQ
5etenv !ALO+KI5H45TMfugue$allmat.dat
"ll the other configuration files must be in the right location.
)nstallation of "-Coffee
2-Coffee is a special mode able to align 2N" se3uences while taking into account
their secondary structure.
Automated )nstallation
&n the T-Coffee distribution, type)
./install rcoffee
&n theory, this command should download and install every re3uired package
#e@cept consan%. &f, however, it fails, you should switch to the manual installation
#see ne@t%.
anual )nstallation
2-Coffee only re3uires the package Gienna to be installed, in order to compute
multiple se3uence alignments. To make the best out of it, you should also have all
the packages re3uired by !-Coffee
Pac&age 7unction
555555555555555555555555555555555555555555555555555
---------------------------------------------------
consan --*offee
*omputes highly accurate pairwise "lignments
18
***.1+ *1#P>?1-@***
selab.'anelia.org/software/consan/
---------------------------------------------------
-."plfold *omputes -." secondary tructures
www.tbi.univie.ac.at/(ivo/-."/
---------------------------------------------------
probcons-." probcons.stanford.edu/
---------------------------------------------------
#-*offee +-*offee and the most common #" Pac&ages
:cf #-*offee in this installation guide<
)nstalling Probbons"9A for "-Coffee
ollow the installation procedure, but make sure you rename the probcons
e@ecutable into probcons2N".
)nstalling Consan for "-Coffee
&n order to insure a proper interface beween consan and 2-Coffee, you must make
sure that the file mi@6-.mod is in the directory N$.tIcoffee$mcoffee or in the mcoffee
directory otherwise declared.
2:
=uic' +tart
/e ol& %ive &o# !'e ve"& $asics 'e"e, -lease #se !'e T#!o"ial 9o"
mo"e de!ailed i9o"ma!io o 'o7 !o #se o#" !ools,
IMPORTANT: All the fles mentionned here (sampe_seq...) can be o!nd in
the e"ample director# o the distrib!tion.
T-C!&&##
'rite your se3uences in the same file #5wiss-prot, asta or (ir% and type.
PROMPT: t_coffee sample_se,A.fasta
This will output two files)
sample_se,A.aln: +our Multiple /e,uence 0lignment
sample_se,A.dnd: T)e :uide tree !ne8icF <ormat#
IMPORTANT:
In theor# n!cleic acids sho!ld be a!tomaticall# detected and the dea!lt
methods sho!ld be adapted appropriatel#. $o%e&er' sometimes this ma#
ail' either beca!se the seq!ences are too short or contain too man#
ambi(!it# codes.
)hen this happens' #o! are ad&ised to e"plicitl# set the t#pe o #o!r
seq!ences
NOT*: the +mode,dna is not needed or s!pported an#more
PROMPT: t_coffee sample_dnase,A.fasta t+pe4dna
-Coffee
!-Coffee is a !eta version of T-Coffee that makes it possible to combine the output
of at least eight packages #!uscle, probcons, poa, dialignT, mafft, clustalw, (C!"
21
and T-Coffee%.
&f all these packages are already installed on your machine. Kou must)
9-set the following environment variables
e"port PO0_$&R4Ga(solute pat) of t)e PO0 installation dirH
e"port $&01&:5T_$&R4G0(solute pat) of t)e $&01&:5-T/conf
Once this is done, write your se3uences in a file and run) same file #5wiss-prot,
asta or (ir% and type.
PROMPT: t_coffee sample_se,A.fasta mode mcoffee
&f the program starts complaining one package or the other is missing, this means
you will have to go the hard way and install all these packages yourself... (roceed to
the !-Coffee section for more detailed instructions.
#-presso
&f you have installed the A4& wublast.pl client, A@presso will 4L"5T your
se3uences against (+4, identify the best targets and use these to align your proteins.
PROMPT: t_coffee sample_se,A.fasta mode e"presso
&f you did not manage to install all the re3uired structural packages for A@presso,
like ugue or 5ap, you can still run e@presso by selecting yourself the structural
packages you want to use. or instance, if youDd rather use T!-"lign than sap, try)
PROMPT: t_coffee sample_se,A.fasta template_file E?PRE//O -met)od
TMalign_pair
"-Coffee
2-Coffee can be used to align 2N" se3uences, using their 2N"pfold predicted
secondary structures. The best results are obtained by using the consan pairwise
method. &f you have consan installed)
t_coffee sample_rnase,A.fasta special_mode rcoffee_consan
22
This will only work if your se3uences are short enough #less than ,-- nucleotides%.
" good alternative is the rmcoffee mode that will run !uscle, (robcons>2N" and
!"fft and then use the secondary structures predicted by 2N"pfold.
PROMPT: t_coffee sample_rnase,A.fasta mode mrcoffee
&f you want to decide yourself which methods should be combined by 2-Coffee,
run)
PROMPT: t_coffee sample_rnase,A.fasta mode rcoffee -met)od
lalign_id_pair slo8_pair
i"+* and AP*%
"ll you need is a file containing the alignment of se3uences with a known structure.
These se3uences must be named according to their (+4 &+, followed by the chain
inde@ # 9aab" for instance%. "ll the se3uences do not need to have a known
structure, but at least two need to have it.
0iven the alignment)
PROMPT: t_coffee ot)er_pg irmsd -aln 3d_sample;.aln
t"+*
t2!5+ is a structure based clustering method using the i2!5+ to drive the
clustering. The T-2!5+ supports all the parameters supported by i2!5+ or
"(+4.
PROMPT: t_coffee ot)er_pg trmsd -aln 3d_sampleB.aln -template_file
3d_sampleB.template_list
8dIsample:.aln is a multiple alignment in which each se3uence has a known
structure. The file 8dIsample:.templateIlist is a fasta like file declaring the
structure associated with each se3uence, in the form)
A Bse!_nameA _P_ BP8= structure file or nameA
23
******* $d_sample).template_list ********
A%>63-$" _P_ %>63-$.pdb
A%>63-%" _P_ %>63-%.pdb
A%>63-C" _P_ %>63-C.pdb
A%/D@-2- _P_ %/D@-2.pdb
...
**************************************
The program then outputs a series of files
+emplate +ype: E$d_sample).template_listF #ode 1r 7ile: E$d_sample).template_listF
EtartF
Eample *olumnsFE+1+5 )CFECGG HFED?"PD8 +3#D: G sec.F
E+ree *mpFE+1+5 C$FE I% HFED?"PD8 +3#D: G sec.F
,,,, 7ile +ype5 +ree?ist 7ormat5 newic& .ame5 $d_sample).tot_pos_list
,,,, 7ile +ype5 +ree 7ormat5 newic& .ame5 $d_sample).struc_treeCG
,,,, 7ile +ype5 +ree 7ormat5 newic& .ame5 $d_sample).struc_tree)G
,,,, 7ile +ype5 +ree 7ormat5 newic& .ame5 $d_sample).struc_treeCGG
,,,, 7ile +ype5 *olored #" 7ormat5 score_html .ame5 $d_sample).struc_tree.html
8dIsample:.totIposIlist is a list of the t2!5+ tree associated with every
position.
8dIsample:.strucItree9-- is a consensus tree #phylip$consense% of the trees
contained in the previous file. This file is the default output
8dIsample:.strucItree9- is a consensus tree #phylip$consense% of the 9-U trees
having the higest average agreement with the rest
8dIsample:.strucItree9- is a consensus tree #phylip$consense% of the :-U trees
having the higest average agreement with the rest
8dIsample:.html is a colored version of the output showing in red the positions
that give the highest support to 8dIsample:.strucItree9--
!CCA
'rite your se3uences in the same file #5wiss-prot, asta or (ir% and type.
PROMPT: t_coffee ot)er_pg mocca sample_se,A.fasta
This command output one files #Pyour se3uencesQ.moccaIlib% and starts an
interactive menu.
2$
"ecent odifications
'arning) This log of recent modifications is not as thorough and accurate as it
should be.
-:.6- Novel assembly algorithm #linkedIpairIwise% and the primary library is now
made of probcons style pairwise alignments #probaIpair%
->.8- and upward) the "V has moved into a new tutorial document
->.8- and upward) -in has will be deprecated and replaced by the flags) -profile,-
method,-aln,-se3,-pdb
->.-,) -modeMdna is still available but not any more needed or supported. Hse
typeMprotein or dna if you need to force things
8.,6) corrected a bug that prevents short se3uences from being correctly aligned
-Hse of * as a separator when specifying methods parameters
-The most notable modifications have to do with the structure of the input. rom
version ,.,-, all files must be tagged to indicate their nature #") alignment, 5)
5e3uence, L) LibraryO%. 'e are becoming stricter, but thatWs for your own goodO
"nother important modification has to do with the flag -matri@) it now controls the
matri@ being used for the computation
2(
"eference anual
This reference manual gives a list of all the flags that can be used to modify the
behavior of T-Coffee. or your convenience, we have grouped them according to
their nature. To display a list of all the flags used in the version of T-Coffee you are
using #along with their default value%, type)
PROMPT: t_coffee
Or
PROMPT: t_coffee )elp
Or
PROMPT: t_coffee )elp in
Or any other parameter
#n5ironment >ariables
I! is possi$le !o modi9& T-Cofee+s $e'avio" $& se!!i% a& o9 !'e
9ollo7i% evi"oeme! va"ia$les, O !'e $as' s'ell, #se e;po"!
<AR=>val#e>, O !'e cs'ell, #se se! ?<AR=>;;;>
http<pro-y<$<TC!&&##
5ets the httpIpro@y and /TT(Ipro@y values used by T-Coffee.
These values get supersede httpIpro@y and /TT(Ipro@y. httpIpro@yI>ITCOAA
2.
gets superseded by the command line values #-pro@y and -email%
&f you have no pro@y, Eust set this value to an empty string.
email<$<TC!&&##
5et the A-mail values provided to web services called upon by T-Coffee. Can be
over-riden by the flag -email.
*)"<$<TC!&&##
4y default this variable is set to R/O!A$.tIcoffee. This is where T-Coffee e@pects
to find its cache, tmp dir and possibly any temporary data stored by the program.
TP<$<TC!&&##
4y default this variable is set to R/O!A$.tIcoffee$tmp. This is where T-Coffee
stores temporary files.
CAC?#<$<TC!&&##
4y default this variable is set to R/O!A$.tIcoffee$cache. This is where T-Coffee
stores any data e@pensive to obtain) pdb files, sap alignments....
P4,@)9+<$<TC!&&##
4y default all the companion packages are searched in the directory
+&2I>ITCOAA$plugins$PO5Q. This variable overrides the default. This variable
can also be overriden by the -plugins T-Coffee flag
9!<#""!"<"#P!"T<$<TC!&&##
4y default this variable is no set. 5et it if you do not want the program to generate a
verbose error output file #useful for running a server%.
P*%<*)"
&ndicate the location of your local (+4 installation.
9!<0A"9)9@<$<TC!&&##
5uppresses all the warnings.
,9)=,#<*)"<$<TC!&&##
5ets)
+&2I>ITCOAA
C"C/AI>ITCOAA
T!(I>ITCOAA
(LH0&N5I>ITCOAA
To the same uni3ue value. The string !H5T be a valid directory
2/
+etting up the T-Coffee en5ironment 5ariables
T-Cofee ca 'ave i!s o7 evi"ome! @le, T'is evi"ome! is 8ep!
i a @le amed ?HO*EA,!5cofeeA!5cofee5ev ad ca $e edi!ed, T'e
val#e o9 a& le%al va"ia$le ca $e modi@ed !'"o#%' !'a! @le, Fo"
is!ace, 'e"e is a e;ample o9 a co@%#"a!io @le 7'e o! "e6#i"i%
a p"o;&,
http_proxy_2_+*177DD5
D#"3?_2_+*177DD5cedric.notredameJeurope.com
IMPORTANT:
-pro"#' -email .. t_co/ee_en& .. en&
0ell %eha5ed Parameters
+eparation
Kou can use any kind of separator you want #i.e. ,7 PspaceQM%. The synta@ used in
this document is meant to be consistent with that of Clustal'. /owever, in order to
take advantage of the automatic filename compleation provided by many shells, you
can replace XMY and X,Y with a space.
Posi-
T-Coffee is not (O5&L compliant.
#ntering the right parameters
There are many ways to enter parameters in T-Coffee, see the -parameter flag in
Para!eters Priority
"n general you #ill not need to use these co!plicated para!eters$ %et, if you find
yourself typing long co!!and lines on a regular basis, it !ay be #orth reading
this section$
&ne !ay easily feel confused #ith the 'arious !anners in #hich the para!eters
can be passed to t(coffee$ The reason for these !any !echanis!s is that they
allo# se'eral le'els of inter'ention$ )or instance, you !ay install t(coffee for all
the users and decide that the defaults #e pro'ide are not the proper ones* "n
this case, you #ill need to !ake your o#n t(coffee(default file$
+ater on, a user !ay find that he,she needs to keep reusing a specific set of
26
para!eters, different fro! those in t(coffee(default, hence the possibility to
#rite an extra para!eter file- para!eters$ "n su!!ary-
para!eters . pro!pt para!eters . t(coffee(defaults . !ode
This !eans that parameters supersede all the others, #hile para!eters pro'ided
'ia special mode are the #eakest$
Parameters +ynta-
No Flag
&f no flag is used <your sequence> must be the first argument. 5ee format for further
information.
PROMPT: t_coffee sample_se,A.fasta
'hich is e3uivalent to
PROMPT: t_coffee /sample_se,A.fasta
'hen you do so, sa!ple(se/0 is used as a name prefi@ for every file the program outputs.
-parameters
1sage- para!eters2para!eters(file
$efault: no parameters file
&ndicates a file containing e@tra parameters. (arameters read this way behave as if they had
been added on the right end of the command line that they either supersede#one value
parameter% or complete #list of values%. or instance, the following file #parameter.file% could
be used
*******sample_param_file.param********
-in5sample_se!C.fasta;#fast_pair
-output5msf_aln
**************************************
3ote- This is one of the exceptions 4#ith 5infile6 #here the identifier tag 47,A,+,M*6
can be o!itted$ Any dataset pro'ided this #ay #ill be assu!ed to be a se/uence 476$
These exceptions ha'e been designed to keep the progra! co!patible #ith Clustal8$
3ote- This para!eter file can &3+% contain 'alid para!eters$ Co!!ents are not
allo#ed$ Para!eters passed this #ay #ill be checked like nor!al para!eters$
Hsed with)
PROMPT: t_coffee -parameters4sample_param_file.param
28
'ill cause tIcoffee to apply the fastIpair method onto to the se3uences contained in
sampleIse3.fasta. &f you wish, you can also pipe these arguments into tIcoffee, by naming
the parameter file .stdin. #as a rule, any file named stdin is e@pected to receive its content
via the stdin%
cat sample_param_file.param I t_coffee -parameters4stdin
-t_coffee_defaults
1sage- t(coffee(defaults29file(na!e.
$efault: not used.
This flag tells the program to use some default parameter file for tIcoffee. The format of that
file is the same as the one used with -parameters. The file used is either)
9. Pfile nameQ if a name has been specified
,. :,$t(coffee(defaults if no file was specified
8. The file indicated by the environment variable TC&));;(<;)A1+T7
-mode
1sage- !ode2 hard coded !ode
$efault: not used.
&t indicates that tIcoffee will use some hard coded parameters. These include)
/uickaln) very fast appro@imate alignment
dali) a mode used to combine dali pairwise alignments
e'aluate) defaults for evaluating an alignment
=dcoffee) runs tIcoffee with the 8dcoffee parameteriFation
Other modes e@ist that are not yet fully supported
-score [Deprecated]
1sage- score
$efault: not used
Toggles on the evaluate mode and causes tIcoffee to evaluates a precomputed alignment
provided via infile29align!ent.. The flag output must be set to an appropriate format
#i.e. -outputMscoreIascii, scoreIhtml or scoreIpdf%. " better default parameteriFation is
obtained when using the flag !ode2e'aluate$
-evaluate
1sage- e'aluate
$efault: not used
2eplaces Zscore. This flag toggles on the evaluate mode and causes tIcoffee to evaluates a
pre-computed alignment provided via infile29align!ent.. The flag output must be set to
an appropriate format #i.e. -outputMscoreIascii, scoreIhtml or scoreIpdf%.
The main purpose of Zevaluate is to let you control every aspect of the evaluation. Ket it is
3:
advisable to use pre-defined parameteriFation) !ode2e'aluate$
PROMPT: t_coffee infile4sample_alnA.aln -mode4ealuate
PROMPT: t_coffee infile4sample_se,A.aln in 1sample_li(A.tc_li(
mode4ealuate
-convert [cw]
1sage- con'ert
$efault: turned off
Toggles on the conversion mode and causes T-Coffee to convert the se3uences, alignments,
libraries or structures provided via the infile and in flags. The output format must be set
via the output flag. This flag can also be used if you simply want to compute a library #i.e.
you have an alignment and you want to turn it into a library%.
This flag is Clustal' compliant.
-do_align [cw]
1sage- do(align
$efault: turned on
+pecial Parameters
-version
1sage- 'ersion
$efault: not used
2eturns the current version number
-proxy
1sage- proxy29proxy.
$efault: not used
5ets the pro@y used by /TT(Ipro@y "N+ httpIpro@y. 5etting with the propmpt supersedes
"NK other setting.
Note that if you use no pro@y, you should set
-pro@y
-email
1sage- e!ail29e!ail.
$efault: not used
5ets your email value as provided to web services
-check_configuration
1sage- check(configuration
31
$efault: not used
Checks your system to determine whether all the programs T-Coffee can interact with are
installed.
-cache
1sage- cache29use, update, ignore, 9filena!e..
$efault: -cac)e4use
4y default, tIcoffee stores in a cache directory, the results of computationally e@pensive
#structural alignment% or network intensive #4L"5T search% operations.
-update
1sage- update
$efault: turned off
Causes a wget access that checks whether the tIcoffee version you are using needs updating.
-full_log
1sage- full(log29filena!e.
$efault: turned off
Causes tIcoffee to output a full log file that contains all the input$output files.
-plugins
1sage- plugins29dir.
$efault: default
5pecifies the directory in which the companion packages #other multiple aligners used by !-
Coffee, structural aligners, etcO% are kept as an alternative, you can also set the environment
variable (LH0&N5I>ITCOAA
The default is N$.tIcoffee$plugins$
-other_pg
1sage- other(pg29filena!e.
$efault: turned off
5ome rumours claim that Tetris is embedded within T-Coffee and could be ran using some
special set of commands. 'e wish to deny these rumours, although we may admit that
several interesting reformatting programs are now embedded in tIcoffee and can be ran
through the ZotherIpg flag.
PROMPT: t_coffee ot)er_pg4se,_reformat
PROMPT: t_coffee ot)er_pg4unpacF_all
PROMPT: t_coffee ot)er_pg4unpacF_e"tract_from_pd(
32
)nput
+e;uence )nput
-infile [cw]
To remain compatible with Clustal', it is possible to indicate the se3uences with this flag
PROMPT: t_coffee -infile4sample_se,A.fasta
3ote- Co!!on !ultiple se/uence align!ents for!at constitute a 'alid input for!at$
3ote- TCoffee auto!atically re!o'es the gaps before doing the align!ent$ This
beha'iour is different fro! that of Clustal8 #here the gaps are kept$
-in !f "in from the #ethod and $ibrary %nput section&
-get_type
1sage- get(type
$efault: turned off
orces tIcoffee to identify the se3uences type #(2OTA&N, +N"%.
-type [cw]
1sage- type2<3A > P?&T;"3> <3A(P?&T;"3
$efault: -t+pe4'automaticall+ set*
This flag sets the type of the se3uences. &f omitted, the type is guessed automatically. This
flag is compatible with Clustal'.
)arnin(: In case o lo% comple"it# or short seq!ences' it is
recommended to set the t#pe man!all#.
-se'
1sage- se/2@9P,7.9na!e.,A
$efault: none
-se3 is now the recommended flag to provide your se3uences. &t behaves mostly like
the -in flag.
-se'_source
1sage- se/(source29A3% or (+7 or +7 .
$efault: 05J.
Kou may not want to combine all the provided se3uences into a single se3uence list. Kou can
do by specifying that you do not want to treat all the Zin files as potential se3uence sources.
-se3IsourceMIL" indicates that neither se3uences provided via the " #"lignment% flag or via
the L #Library flag% should be added to the se3uence list.
-se3IsourceM5 means that only se3uences provided via the 5 tag will be considered. "ll the
33
other se3uences will be ignored.
3ote- This flag is !ostly designed for interactions bet#een TCoffee and TCoffee<PA
4the large scale 'ersion of TCoffee6$
+tructure )nput
-pdb
1sage- pdb29pdbid0.,9pdbidB.*@Max BCCA
$efault: 5one
2eads or fetch a pdb file. &t is possible to specify a chain or even a sub-chain)
P$%&$!P$%_6H0&5#GoptH !<&R/TK10/T#GoptH
&t is also possible to input structures via the Zin flag. &n that case, you will need to use the
T"0 identifier)
-in Ppd(A Ppd(DL
Tree )nput
-usetree
1sage- usetree29tree file.
$efault: 5o file specified
<ormat: ne8icF tree format !6lustal= /t+le#
This flag indicates that rather than computing a new dendrogram, tIcoffee must use a pre-
computed one. The tree files are in phylips format and compatible with Clustal'. &n most
cases, using a pre-computed tree will halve the computation time re3uired by tIcoffee. &t is
also possible to use trees output by Clustal', (hylips and any other program.
+tructures3 +e;uences ethods and 4ibrary )nput
5ia the Ain &lag
The in )lag and its "dentifier TAD7
9in. is the real grinder of TCoffee$ 7e/uences, !ethods and align!ents all pass
through so that TCoffee can turn it all into a single list of constraints 4the
library6$ ;'erything is done auto!atically #ith TCoffee going through each file
to extract the se/uences it contains$ The !ethods are then applied to the
se/uences$ Preco!piled constraint list can also be pro'ided$ ;ach file pro'ided
'ia this flag !ust be preceded #ith a sy!bol 4"dentifier TAD6 that indicates its
nature to TCoffee$ The TADs currently supported are the follo#ing-
P P<B structure
7 for se/uences 4use it as #ell to treat an M7A as unaligned se/uences6
3$
M Methods used to build the library
+ Preco!puted TCoffee library
A Multiple Align!ents that !ust be turned into a +ibrary
E 7ubstitution !atrices$
? Profiles$ This is a legal !ultiple align!ents that #ill be treated as single
se/uences 4the se/uences it contains #ill not be realigned6$
"f you do not #ant to use the TAD7, you #ill need to use the follo#ing flags in
replace!ent of in$ <o not use the TAD7 #hen using these flags-
aln Align!ents 4A6
profile Profiles 4?6
!ethod Method 4M6
se/ 7e/uences 476
lib +ibraries 4+6
-in
1sage- in2@9P,7,A,+,M,E.9na!e.,A
$efault: -in4Mlalign_id_pairKMclustal8_pair
Note: -in can be replaced %ith the combined !sa(e o -aln' iprofle'
.pdb' .lib' -method.
5ee the bo@ for an e@planation of the -in flag. The following argument passed via -in
PROMPT: t_coffee
-in4/sample_se,A.fastaK0sample_alnA.alnK0sample_alnD.msfKMlalign_i
d_pairK1sample_li(A.tc_li( outfile4outaln
This command will trigger the following chain of events)
9-0ather all the se3uences
5e3uences within all the provided files are pooled together. ormat recognition is automatic.
+uplicates are removed #if they have the same name%. +uplicates in a single file are only
tolerated in "5T" format file, although they will cause se3uences to be renamed.
&n the above case, the total set of se3uences will be made of se3uences contained in
se3uences9.se3, alignment9.aln, alignment,.msf and library.lib, plus the se3uences initially
gathered by -infile.
,-Turn alignments into libraries
alignment9.aln and alignment,.msf will be read and turned into libraries. "nother library
will be produced by applying the method lalignIidIpair to the set of se3uences previously
obtained #9%. The final library used for the alignment will be the combination of all this
information.
Note as well the following rules)
3(
0&rder) The order in which se3uences, methods, alignments and libraries are fed in is
irrelevant.
BHeterogeneity) There is no need for each element #", 5, L% to contain the same se3uences.
=3o <uplicate) Aach file should contain only one copy of each se3uence. +uplicates are
only allowed in "5T" files but will cause the se3uences to be renamed.
F?econciliation) &f two files #for instance two alignments% contain different versions of the
same se3uence due to an indel, a new se3uence will be reconstructed and used instead)
aln A:)ga(A 00000%00000
aln D:)ga(A 0000000000666
will cause the program to reconstruct and use the following se3uence
)ga(A 00000%00000666
This can be useful if you are trying to combine several runs of blast, or structural
information where residues may have been deleted. /owever substitutions are forbidden. &f
two se3uences with the same name cannot be merged, they will cause the program to e@it
with an information message.
GMethods) The method describer can either be built in #5ee [[[ for a list of all the
available methods% or be a file describing the method to be used. The e@act synta@ is
provided in part > of this manual.
H7ubstitution Matrices) &f the method is a substitution matri@ #L% then no other type of
information should be provided. or instance)
PROMPT: t_coffee sample_se,A.fasta -in4?pamDB2mt -gapopen4-A2
-gape"t4-A
This command results in a progressive alignment carried out on the se3uences in se3file. The
procedure does not use any more the T-Coffee concistency based algorithm, but switches to
a standard progressive alignment algorithm #like Clustal' or (ileup% much less accurate. &n
this conte@t, appropriate gap penalties should be provided. The matrices are in the file
source$matrices.h. "dd-/oc matrices can also be provided by the user #see the matrices
format section at the end of this manual%.
)arnin(: 0matri" does not ha&e the same e/ect as !sin( the -matri" 1a(.
The -matri" defnes the matri" that %ill be !sed %hile compilin( the
librar# %hile the 0matri" defnes the matri" !sed %hen assemblin( the
fnal ali(nment.
Profile )nput
-profile
1sage- profile2@9na!e.,A !axi!u! of BCC profiles$
$efault: no default
This flag causes T-Coffee to treat multiple alignments as a single se3uences, thus making it
possible to make multiple profile alignments. The profile-profile alignment is controlled by
-profileImode and -profileIcomparison. 'hen provided with the in flag, profiles must be
3.
preceded with the letter 2.
PROMPT: t_coffee profile sample_alnA.alnKsample_alnD.aln
outfile4profile_aln
PROMPT: t_coffee in
Rsample_alnA.alnKRsample_alnD.alnKMslo8_pairKMlalign_id_pair
outfile4profile_aln
Note that when using ZtemplateIfile, the program will also look for the templates associated
with the profiles, even if the profiles have been provided as templates themselves #however
it will not look for the template of the profile templates of the profile templatesO%
-profile( [cw]
1sage- profile02@9na!e.A, one na!e only
$efault: no default
5imilar to the previous one and was provided for compatibility with Clustal'.
-profile) [cw]
1sage- profile02@9na!e.A, one na!e only
$efault: no default
5imilar to the previous one and was provided for compatibility with Clustal'.
Alignment Computation
4ibrary Computation: ethods
-lalign_n_top
1sage- lalign(n(top29"nteger.
$efault: -lalign_n_top4A2
Number of alignment reported by the local method #lalign%.
-align_pdb_param_file
Hnsuported
-align_pdb_hasch_mode
Hnsuported
4ibrary Computation: #-tension
-lib_list [*nsupported]
1sage- lib(list29filena!e.
$efault:unset
Hse this flag if you do not want the library computation to take into account all the possible
pairs in your dataset. or instance
3/
ormat)
% .ameC name%
% .ameC name2
$ .ameC .ame% .ame$K
#the line 8 would be used by a multiple alignment method%.
-do_normalise
1sage- do(nor!alise29C or a positi'e 'alue.
$efault:-do_normalise4A222
$eelopment Onl+
'hen using a value different from -, this flag sets the score of the highest scoring pair to
9---.
-extend
1sage- extend29C,0 or a positi'e 'alue.
$efault:-e"tend4A
$eelopment Onl+
'hen turned on, this flag indicates that the library e@tension should be carried out when
performing the multiple alignment. &f extend 2C, the e@tension is not made, if it is set to 9,
the e@tension is made on all the pairs in the library. &f the e@tension is set to another positive
value, the e@tension is only carried out on pairs having a weight value superior to the
specified limit.
-extend_mode
1sage- extend29string.
$efault:-e"tend4er+_fast_triplet
=arning: $eelopment Onl+
Controls the algorithm for matri@ e@tension. "vailable modes include)
relativeItriplet Hnsupported
gIcoffee Hnsupported
gIcoffeeI3uadruplets Hnsupported
fastItriplet ast triplet e@tension
veryIfastItriplet slow triplet e@tension, limited to the !ax(n(pair best
se3uence pairs when aligning two profiles
slowItriplet A@haustive use of all the triplets
mi@t Hnsupported
3uadruplet Hnsupported
test Hnsupported
matri@ Hse of the matri@ !atrix
fastImatri@ Hse of the matri@ !atrix. (rofiles are turned into consensus
36
-max_n_pair
1sage- !ax(n(pair29integer.
$efault:-e"tend4A2
$eelopment Onl+
Controls the number of pairs considered by the extend(!odeMveryIfastItriplet. 5etting it
to - forces all the pairs to be considered #e3uivalent to extend(!odeMslowItriplet%.
-se'_name_for_'uadruplet
1sage- 1nsupported
-compact
1sage- 1nsupported
-clean
1sage- 1nsupported
-maximise
1sage- 1nsupported
-do_self
1sage- )lag do(self
<efault- 3o
This flag causes the e@tension to carried out within the se3uences #as opposed to between
se3uences%. This is necessary when looking for internal repeats with !occa.
-se'_name_for_'uadruplet
1sage- 1nsupported
-weight
1sage- #eight29#insi!3, si! or si!(9!atrix(na!e or
!atrix(file. or 9integer 'alue.
$efault: -8eig)t4sim
'eight defines the way alignments are weighted when turned into a library. Overweighting
can be obtained with the O'PLQ weight mode.
winsimN indicates that the weight assigned to a given pair will be e3ual to the percent
identity within a window of ,N\9 length centered on that pair. or instance winsim9-
defines a window of 9- residues around the pair being considered. This gives its own weight
to each residue in the output library. &n our hands, this type of weighting scheme has not
provided any significant improvement over the standard sim value.
PROMPT: t_coffee sample_se,A.fasta -8eig)t48insimA2
out_li(4test.tc_li(
sim indicates that the weight e3uals the average identity within the se3uences containing the
38
matched residues.
+,-./ 'ill cause the sim weight to be multiplied by L
sim_matrix_name indicates the average identity with two residues regarded as identical
when their substitution value is positive. The valid matrices names are in matrices.h
(pam250mt) .!atrices not found in this header are considered to be filenames. 5ee the
format section for matrices. or instance, -weight=sim_pam250mt indicates that the
grouping used for similarity will be the set of classes with positive substitutions.
PROMPT: t_coffee sample_se,A.fasta -8eig)t48insimA2
out_li(4test.tc_li(
Other groups include
sim_clustalw_col # categories of clustalw marked with )%
sim_clustalw_dot # categories of clustalw marked with .%
0alue indicates that all the pairs found in the alignments must be given the same weight
e3ual to value. This is useful when the alignment one wishes to turn into a library must be
given a pre-specified score #for instance if they come from a structure super-imposition
program%. Galue is an integer)
PROMPT: t_coffee sample_se,A.fasta -8eig)t4A222
out_li(4test.tc_li(
Tree Computation
-distance_matrix_mode
1sage- distance(!atrix(!ode29slo#, fast, 'ery(fast.
$efault: er+_fast
This flag indicates the method used for computing the distance matri@ #distance between
every pair of se3uences% re3uired for the computation of the dendrogram.
7lo# The chosen dpImode using the e@tended library,
fast) The fasta dpImode using the e@tended library.
'ery(fast The fasta dpImode using blosum=,mt.
ktup Ctup matching #!uscle kind%
aln 2ead the distances on a precomputed !5"
-'uicktree [!,]
1sage- /uicktree
$escription: 6auses T-6offee to compute a fast appro"imate
guide tree
This flag is kept for compatibility with Clustal'. &t indicates that)
PROMPT: t_coffee sample_se,A.fasta distance_matri"_mode4er+_fast
$:
PROMPT: t_coffee sample_se,A.fasta ,uicFtree
Pair-1ise Alignment Computation
Controlling Align!ent Co!putation
Most para!eters in this section refer to the align!ent !ode fasta(pair(#ise and
cfatsa(pair(#ise$ 8hen using these align!ent !odes, things proceed as follo#-
07e/uences are recoded using a degenerated alphabet pro'ided #ith 9
si!(!atrix.
B?ecoded se/uences are then hashed into ktuples of siIe 9ktup.
=<yna!ic progra!!ing runs on the 9ndiag. best diagonals #hose score is
higher than 9diag(threshold., the #ay diagonals are scored is controlled 'ia 9
diag(!ode. $
FThe <yna!ic co!putation is !ade to opti!iIe either the library scoring
sche!e 4as defined by the in flag6 or a substitution !atrix as pro'ided 'ia the
!atrix flag$ The penalty sche!e is defined by gapopen and gapext$ "f gapopen
is undefined, the 'alue defined in cos!etic(penalty is used instead$
GTer!inal gaps are scored according to tg(!ode
-dp_mode
1sage- dp(!ode29string.
$efault: -dp_mode4cfasta_fair_8ise
This flag indicates the type of dynamic programming used by the program)
PROMPT: t_coffee sample_se,A.fasta dp_mode m+ers_miller_pair_8ise
gotoh_pair_wise) implementation of the gotoh algorithm #3uadratic in memory and time%
myers_miller_pair_wise) implementation of the !yers and !iller dynamic programming
algorithm # 3uadratic in time and linear in space%. This algorithm is recommended for very
long se3uences. &t is about , times slower than gotoh and only accepts tg_mode=or 2 (i.e.
gaps penali!ed "or opening).
fasta_pair_wise# implementation of the fasta algorithm. The se3uence is hashed, looking for
$tuples words. +ynamic programming is only carried out on the ndiag best scoring
diagonals. This is much faster but less accurate than the two previous. This mode is
controlled by the parameters ktuple, diag(!ode and ndiag
cfasta_pair_wise) c stands for checked. &t is the same algorithm. The dynamic programming
is made on the ndiag best diagonals, and then on the ,Jndiags, and so on until the scores
converge. Comple@ity will depend on the level of divergence of the se3uences, but will
usually be LJlog#L%, with an accuracy comparable to the two first mode # this was checked
on 4ali4ase%. This mode is controlled by the parameters ktuple, diag(!ode and 5ndiag
3ote- 1sers !ay find by looking into the code that other !odes #ith fancy na!es exists
4'iterby(pair(#ise*6 1nless !entioned in this docu!entation, these !odes are not
$1
supported$
-ktuple
1sage- ktuple29'alue.
$efault: -Ftuple4A or D
&ndicates the ktuple siFe for cfastaIpairIwise dpImode and fastaIpairIwise. &t is set to 9 for
proteins, and , for +N". The alphabet used for protein can be a degenerated version, set
with si!(!atrix$.
-ndiag
1sage- ndiag29'alue.
$efault: -ndiag42
&ndicates the number of diagonals used by the "asta_pair_wise algorithm #cf dp(!ode%.
'hen ndiag2C, nIdiagMLog #length of the smallest se3uence%\9.
8hen 5ndiag and 5diag(threshold are set, diagonals are selected if and only if they
fulfill both conditions$
-diag_mode
1sage- diag(!ode29'alue.
$efault: -diag_mode42
&ndicates the manner in which diagonals are scored during the fasta hashing.
-) indicates that the score of a diagonal is e3ual to the sum of the scores of the e@act matches
it contains.
9 indicates that this score is set e3ual to the score of the best uninterrupted segment #useful
when dealing with fragments of se3uences%.
-diag_threshold
1sage- diag(threshold29'alue.
$efault: -diag_t)res)old42
5ets the value of the threshold when selecting diagonals.
-) indicates that Zndiag should be used to select the diagonals #cf Zndiag section%.
-sim_matrix
1sage- si!(!atrix29string.
$efault: -sim_matri"4asiliF+
&ndicates the manner in which the amino acid alphabet is degenerated when hashing in the
fastaIpairwise dynamic programming. 5tandard Clustal' matrices are all valid. They are
used to define groups of amino acids having positive substitution values. &n T-Coffee, the
default is a 98 letter grouping named Gasiliky, with residues grouped as follows)
rFK deK ,)K ilmK f+ !ot)er residues Fept alone#.
This alphabet is set with the flag si!(!atrix2'asiliky. &n order to keep the alphabet non
degenerated, si!(!atrix2id!at can be used to retain the standard alphabet.
$2
-matrix [!,]
1sage- !atrix29blosu!HB!t.
$efault: -matri"4(losum3Dmt
The usage of this flag has been modified from previous versions, due to fre3uent mistakes in
its usage. This flag sets the matri@ that will be used by alignment methods within tIcoffee
#slowIpair, lalignIidIpair%. &t does not affect e@ternal methods #like clustalIpair,
clustalIalnO%.
Hsers can also provide their own matrices, using the matri@ format described in the
appendi@.
-nomatch
1sage- no!atch29positi'e 'alue.
$efault: -nomatc)42
&ndicates the penalty to associate with a match. 'hen using a library, all matches are
positive or e3ual to -. !atches e3ual to - are unsupported by the library but non-penaliFed.
5etting nomatch to a non-negative value makes it possible to penaliFe these null matches and
prevent unrelated se3uences from being aligned #this can be useful when the alignments are
meant to be used for structural modeling%.
-gapopen
1sage- gapopen29negati'e 'alue.
$efault: -gapopen42
&ndicates the penalty applied for opening a gap. The penalty must be negative. &f no value is
provided when using a substitution matri@, a value will be automatically computed.
/ere are some guidelines regarding the tuning of gapopen and gape@t. &n T-Coffee matches
get a score between - #match% and 9--- #match perfectly consistent with the library%. The
default cosmetic penalty is set to -:- #:U of a perfect match%. &f you want to tune -gapoen
and see a strong effect, you should therefore consider values between - and -9---.
-gapext
1sage- gapext29negati'e 'alue.
$efault: -gape"t42
&ndicates the penalty applied for e@tending a gap #cf gapopen%
-fgapopen
1nsupported
-fgapext
1nsupported
-cosmetic_penalty
1sage- cos!etic(penalty29negati'e 'alue.
$efault: -cosmetic_penalt+4-B2
&ndicates the penalty applied for opening a gap. This penalty is set to a very low value. &t
will only have an influence on the portions of the alignment that are unalignable. &t will not
$3
make them more correct, but only more pleasing to the eye # i.e. "void stretches of lonely
residues%.
The cosmetic penalty is automatically turned off if a substitution matri@ is used rather than a
library.
-tg_mode
1sage- tg(!ode29C, 0, or B.
$efault: -tg_mode4A
-) terminal gaps penaliFed with -gapopen 1 -gapext2len
9) terminal gaps penaliFed with a -gapext2len
,) terminal gaps unpenaliFed.
0eighting +chemes
-se'_weight
1sage- se/(#eight29t(coffee or 9file(na!e..
$efault: -se,_8eig)t4t_coffee
These are the individual weights assigned to each se3uence. The tIcoffee weights try to
compensate the bias in consistency caused by redundancy in the se3uences.
sim#",4%MUsimilarity between " and 4, between - and 9.
weight#"%M9$sum#sim#",L%]8%
'eights are normaliFed so that their sum e3uals the number of se3uences. They are applied
onto the primary library in the following manner)
resIscore#"@,4y%M!in#weight#"%, weight#4%%JresIscore#"@, 4y%
These are very simple weights. Their main goal is to prevent a single se3uence present in
many copies to dominate the alignment.
3ote- The library output by out(lib is the un#eighted library$
3ote- 8eights can be output using the outse/#eight flag$
3ote- %ou can use your o#n #eights 4see the for!at section6$
ultiple Alignment Computation
-msa_mode
1sage- !sa(!ode29tree,graph,preco!puted.
$efault: -ealuate_mode4tree
Hnsupported
-one)all
1sage- oneBall29na!e.
$$
$efault: not used
'ill generate a one to all library with respect to the specified se3uence and will then align
all the se3uences in turn to that se3uence, in a se3uence determined by the order in which the
se3uences were provided.
5profile(co!parison 2profile, the !5"s provided via Zprofile are vectoriFed and the
function specified by ZprofileIcomparison is used to make profile profile alignments. &n that
case, the comple@ity is NL],
-profile_comparison
1sage- profile(!ode29full3,profile.
$efault: -profile_mode4fullB2
The profile mode flag controls the multiple profile alignments in T-Coffee. There are two
instances where tIcoffee can make multiple profile alignments)
9-'hen N, the number of se3uences is higher than 5!axnse/, the program switches to its
multiple profile alignment mode #tIcoffeeIdpa%.
,-'hen !5"s are provided via the 5profile flag or via 5profile0 and 5profileB.
&n these situations, the ZprofileImode value influences the alignment computation, these
values are)
5profile(co!parison 2profile, the !5"s provided via Zprofile are vectoriFed and the
function specified by ZprofileIcomparison is used to make profile profile alignments. &n that
case, the comple@ity is NL],
-profle_comparison,!llN, N is an integer value that can omitted. %ull
indicates that given two profiles, the alignment will be based on a library that includes every
possible pair of se3uences between the two profiles. &f N is set, then the library will be
restricted to the N most similar pairs of se3uences between the two profiles, as Eudged from
a measure made on a pairwise alignment of these two profiles.
-profile_mode
1sage- profile(!ode29c#(profile(profile, !uscle(profile(profile,
!ulti(channel.
$efault: -profile_mode4c8_profile_profile
'hen 5profile(co!parison2profile, this flag selects a profile scoring function.
Alignment Post-Processing
-clean_aln
1sage- clean(aln
$efault:-clean_aln
This flag causes T-Coffee to post-process the multiple alignment. 2esidues that have a
reliability score smaller or e3ual to -cleanIthreshold #as given by an evaluation that uses
-cleanIevaluateImode% are realigned to the rest of the alignment. 2esidues with a score
higher than the threshold constitute a rigid framework that cannot be altered.
The cleaning algorithm is greedy. &t starts from the top left segment of low constituency
residues and works its way left to right, top to bottom along the alignment. Kou can re3uire
this operation to be carried out for several cycles using the -cleanIiterations flag.
The rationale behind this operation is mostly cosmetic. &n order to ensure a decent looking
$(
alignment, the gop is set to -,- and the gep to -9. There is no penalty for terminal gaps, and
the matri@ is blosum=,mt.
3ote- Daps are al#ays considered to ha'e a reliability score of C$
Note: The !se o the cleanin( option can res!lt in memor#
o&er1o% %hen ali(nin( lar(e seq!ences'
-clean_threshold
1sage- clean(threshold29CJ.
<efault-clean(aln20
5ee -cleanIaln for details.
-clean_iteration
1sage- clean(iteration29'alue bet#een 0 and .
$efault:-clean_iteration4A
5ee -cleanIaln for details.
-clean_evaluation_mode
1sage- clean(iteration29e'aluation(!ode .
$efault:-clean_iteration4t_coffee_non_e"tended
&ndicates the mode used for the evaluation that will indicate the segments that should be
realigned. 5ee -evaluationImode for the list of accepted modes.
-iterate
1sage- iterate29integer.
$efault: -iterate42
5e3uences are e@tracted in turn and realigned to the !5". &f iterate is set to -9, each
se3uence is realigned, otherwise the number of iterations is set by Ziterate.
CP, Control
ultithreading
-multi_core
1sage- !ulti(core2 te!plates(Kobs(relax(!sa
$efault: 2
template: fetc) t)e templates in a parallel 8a+
Mo(s: compute t)e li(rar+
rela": e"tend t)e li(rar+ in a parallel 8a+
msa: compute t)e msa in a parallel 8a+
5pecifies that the steps of T-Coffee that should be multi threaded. by default all relevant
$.
steps are paralleliFed.
PROMPT: t_coffee sample_se,D.fasta -multi_core Mo(s
&n order to prevent the use of the parallel mode it is possible to use)
PROMPT: t_coffee sample_se,D.fasta -multi_core no
-n_core
1sage- n(core2 9nu!ber of cores.
$efault: 2
+efault indicates that all cores will be used, as indicated by the environment via)
PROMPT: t_coffee sample_se,D.fasta -multi_core Mo(s
4imits
-mem_mode
1sage- deprecated
-ulimit
1sage- uli!it29'alue.
$efault: -ulimit42
5pecifies the upper limit of memory usage #in !egabytes%. (rocesses e@ceeding this limit
will automatically e@it. " value - indicates that no limit applies.
-maxlen
1sage- !axlen29'alue, C2noli!it.
$efault: -ma"len4A222
&ndicates the ma@imum length of the se3uences.
Aligning more than 1:: se;uences 1ith *PA
-maxnse'
1sage- !axnse/29'alue, C2noli!it.
$efault: -ma"nse,4B2
&ndicates the ma@imum number of se3uences before triggering the use of tIcoffeeIdpa.
$/
-dpa_master_aln
1sage- dpa(!aster(aln29)ile, !ethod.
$efault: -dpa_master_aln45O
'hen using dpa, tIcoffee needs a seed alignment that can be computed using any
appropriate method. 4y default, tIcoffee computes a fast appro@imate alignment.
" pre-alignment can be provided through this flag, as well as any program using the
following synta@)
+our_script in 'fasta_file* -out 'file_name*
-dpa_maxnse'
1sage- dpa(!axnse/29integer 'alue.
$efault: -dpa_ma"nse,432
!a@imum number of se3uences aligned simultaneously when +(" is ran. 0iven the tree
computed from the master alignment, a node is sent to computation if it controls more than 5
dpa(!axnse/ O2 if it controls a pair of se3uences having less than 5dpa(!in(scoreB
percent &+.
-dpa_min_score(
1sage- dpa(!in(score029integer 'alue.
$efault: -dpa_min_scoreA4EB
Threshold for not realigning the se3uences within the master alignment. 0iven this
alignment and the associated tree, se3uences below a node are not realigned if none of them
has less than 5dpa(!in(score0 U identity.
-dpa_min_score)
1sage- dpa(!in(scoreB
$efault: -dpa_min_scoreD
!a@imum number of se3uences aligned simultaneously when +(" is ran. 0iven the tree
computed from the master alignment, a node is sent to computation if it controls more than 5
dpa(!axnse/ O2 if it controls a pair of se3uences having less than 5dpa(!in(scoreB
percent &+.
-dap_tree [N+3 %#4$5#5N35D]
1sage- dpa(tree29filena!e.
$efault: -unset
0uide tree used in +(". This is a newick tree where the distance associated with each node
is set to the minimum pairwise distance among all considered se3uences.
$6
,sing +tructures
@eneric
-mode
1sage- !ode2=dcoffee
$efault: turned off
2uns tIcoffee with the 8dcoffee mode #cf ne@t section%.
-check_pdb_status
1sage- check(pdb(status
$efault: turned off
orces tIcoffee to run e@tractIfromIpdb to check the pdb status of each se3uence. This can
considerably slow down the program.
3* Coffee: ,sing +AP
&t is possible to use tIcoffee to compute multiple structural alignments. To do so, ensure that
you have the sap program installed.
PROMPT: t_coffee pd(4strucA.pd(KstrucD.pd(Kstruc3.pd( -met)od
sap_pair
'ill combine the pairwise alignments produced by 5"(. There are currently four methods
that can be interfaced with tIcoffee)
sapIpair) that uses the sap algorithm
alignIpdb) uses a tIcoffee implementation of sap, not as accurate.
tmaliagnIpair #http)$$Fhang.bioinformatics.ku.edu$T!-align$%
mustangIpair #http)$$www.cs.mu.oF.au$Narun$mustang%
'hen providing a (+4 file, the computation is only carried out on the first chain of this file.
&f your original file contains several chain, you should e@tract the chain you want to work
on. Kou can use t(coffee 5other(pg extract(fro!(pdb or any pdb handling program.
&f you are working with public (+4 files, you can use the (+4 identifier and specify the
chain by adding its inde@ to the identifier #i.e. 9pdbC%. &f your structure is an N!2 structure,
you are advised to provide the program with one structure only.
&f you wish to align only a portion of the structure, you should e@tract it yourself from the
pdb file, using t(coffee 5other(pg extract(fro!(pdb or any pdb handling program.
Kou can provide tIcoffee with a mi@ture of se3uences and structure. &n this case, you should
use the special mode)
PROMPT: t_coffee mode 3dcoffee se, 3d_sample3.fasta
-template_file template_file.template
$8
,sing/finding P*% templates for the +e;uences
-template_file
1sage- te!plate(file 2
9filena!e,
7C?"PT(scripta!e,
7;+)(TAD
7;L)"+;(TAD(filena!e,
no.
$efault: no
This flag instructs tIcoffee on the templates that will be used when combining several types
of information. or instance, when using structural information, this file will indicate the
structural template that corresponds to your se3uences. The identifier T indicates that the file
should be a "5T" like file, formatted as follows. There are several ways to pass the
templates)
4redefined #odes
AL(2A55O) will use the A4& server to find I(I templates
(5&4L"5T) will use the A4& sever to find profiles
File name
This file contains the se3uence$template association it uses a "5T"-like format, as follows)
ABse!uence nameA _P_ Bpdb templateA
ABse!uence nameA _0_ Bgene templateA
ABse!uence nameA _-_ B#" templateA
ABse!uence nameA _7_ B-." econdary tructureA
ABse!uence nameA _+_ B+ransmembrane econdary tructureA
ABse!uence nameA _D_ BProtein econdary tructureA
Aach template will be used in place of the se3uence with the appropriate method. or
instance, structural templates will be aligned with sapIpair and the information thus
generated will be transferred onto the alignment.
Note the following rule)
-Aach se3uence can have one template of each type #structural, genomicsO%
-Aach se3uence can only have one template of a given type
-5everal se3uences can share the same template
-"ll the se3uences do not need to have a template
The type of template on which a method works is declared with the 5AVITK(A parameter
in the method configuration file)
5AVITK(A 5) a method that uses se3uences
5AVITK(A (5) a pairwise method that aligns se3uences and structures
5AVITK(A () a method that aligns structures #sap for instance%
There are > tags identifying the template type)
I(I 5tructural templates) a pdb identifier O2 a pdb file
I0I 0enomic templates) a protein se3uence where boundary amino-acid have been
(:
recoded with # o)-, i)9, E),%
I2I (rofile Templates) a file containing a multiple se3uence alignment
II 2N" secondary 5tructures
!ore than one template file can be provided. There is no need to have one template for
every se3uence in the dataset.
I(I, I0I, and I2I are known as te!plate TAD7
)-6!7%43_-scriptname/
&ndicates that filename is a script that will be used to generate a valid template file. The
script will run on a file containing all your se3uences using the following synta@)
scriptname infile4'+our se,uences*
-outfile4'template_file*
&t is also possible to pass some parameters, use * as a separator and [ in place of the M sign.
or instance, if you want to call the a script named blast.pl with the foloowing parameters7
(last.pl -d(4pd( -dir4/local/test
Hse
/6R&PT_(last.plNd(Opd(NdirO/local/test
4ear in mind that the input output flags will then be concatenated to this command line so
that tIcoffee ends up calling the program using the following system call)
(last.pl -d(4pd( -dir4/local/test -infile4'some tmp file*
-outfile4'anot)er tmp file*
8-65$F_39:
T"0 can take the value of any of the known T"05 #I5I, I0I, I(I%. 5AL indicates that the
original name of the se3uence will be used to fetch the template)
PROMPT: t_coffee 3d_sampleD.fasta template_file /E1<_P_
The previous command will work because the se3uences in 8dIsample8 are named
;-65<F%$5_39:_filename
Hse this flag if your templates are in filename, and are named according to the se3uences.
or instance, if your protein se3uences have been recoded with A@on$&ntron information, you
should have the recoded se3uences names according to the original)
(1
/EP<&1E_:_recodedprotein.fasta
-struc_to_use
1sage- struc(to(use29struc0, strucB*.
$efault: -struc_to_use45911
2estricts the 8+coffee to a set of pre-defined structures.
ultiple 4ocal Alignments
&t is possible to compute multiple local alignments, using the moca routine. !OC"
is a routine that allows e@tracting all the local alignments that show some similarity
with another predefined fragment.
DmoccaD is a perl script that calls t-coffee and provides it with the appropriate
parameters.
-domain=-mocca
1sage- do!ain
$efault: not set
This flag indicates that tIcoffee will run using the domain mode. "ll the se3uences will be
concatenated, and the resulting se3uence will be compared to itself using lalignIrsIsIpair
mode #lalign of the se3uence against itself using keeping the lalign raw score%. This step is
the most computer intensive, and it is advisable to save the resulting file.
PROMPT: t_coffee -in /sample_se,A.fastaKMlalign_rs_s_pair
-out_li(4sample_li(A.mocca_li( -domain -start4A22 -len4B2
This instruction will use the fragment 9---9:- on the concatenated se3uences, as a template
for the e@tracted repeats. The e@traction will only be made once. The library will be placed
in the file Plib nameQ.
&f you want, you can test other coordinates for the repeat, such as
PROMPT: t_coffee -in sample_li(A.mocca_li( -domain -start4A22
-len432
This run will use the fragment 9---9=-, and will be much faster because it does not need to
re-compute the lalign library.
-start
1sage- start29int 'alue.
$efault: not set
This flag indicates the starting position of the portion of se3uence that will be used as a
template for the repeat e@traction. The value assumes that all the se3uences have been
concatenated, and is given on the resulting se3uence.
(2
-len
1sage- len29int 'alue.
$efault: not set
This flag indicates the length of the portion of se3uence that will be used as a template.
-scale
1sage- scale29int 'alue.
$efault: -scale4-A22
This flag indicates the value of the threshold for e@tracting the repeats. The actual threshold
is e3ual to)
motifIlenJscale
&ncrease the scale &ncrease sensitivity !ore alignments# i.e. -:-%.
-domain_interactive [5xamples]
1sage- do!ain(interacti'e
$efault: unset
Launches an interactive mocca session.
PROMPT: t_coffee -in 1sample_li(3.tc_li(KMlalign_rs_s_pair -domain
-start4A22 -len432
+1?=_D*1?3_%C%_%L %CC M?"@4+7D0---"?43N+?".0"4-N4-
"7P-/.0"P"7P80M?"7"
+1?=_D*1?3_CL)_%CO CL2 +-3"@44N+.00N7P@D?-48@80@.N744/-PNP?#P"6P80M?"@4
+1?=_D*1?3_%)L_$GL %)) M?"7"?M+0--?.?@4#8?"0N3-N4-+80-..+DP+67P8N.?"7+
+1?=_D*1?3_$GP_$)G $GL -------8N"0---PN4@M4.3.00"PN-3-+6D0N.N8"8480M7#4#4
+1?=_D*1?3_$)C_$I$ $)G -------.00N--N/3"MN8?"+004-N4-?+7?8D+P?"P.0+#43@
C * * : . .:. :
#D.>: +ype ?etter 7lagEnumberF and -eturn: ex QCG
Qx --Aet the +"-+ to x
Ax --Aet the ?D. to x
*x --Aet the s*ale to x
name --Aave the "lignment
=x --Aave 0oes bac& x it
return --A*ompute the "lignment
9 --Ae9it
E3+D-"+31. CF E+"-+5%CCF E?D.5 )GF E*"?D5-CGGF @1>- */13*D:
7or instance; to set the length of the domain to 2G; type:
E3+D-"+31. CF E+"-+5%CCF E?D.5 )GF E*"?D5-CGGF @1>- */13*D:A2GEreturnF
EreturnF
6hich will generate:
+1?=_D*1?3_%C%_%)% %CC M?"@4+7D0-"?43N+?".0"4-N4"7P-/.0"P"7 %)C
+1?=_D*1?3_%)L_%IL %)) M?"7"?M+0?.?@4#8?"0N3-N4+80-..+DP+6 %I)
+1?=_D*1?3_$GG_$2G %II N.?"7+8N"0-PN4@M4.3.00"PN-3+6D0N.N8"84 $$I
+1?=_D*1?3_$22_$O$ $2$ M7#4#4.00NN/3"MN8?"+004-N4?+7?8D+P? $O%
+1?=_D*1?3_$OP_2%P $OL +#43@N0#04?.?4+80-7M"-?P"+80N4M7P"6 2%L
C : : : :: . 2G
#D.>: +ype ?etter 7lagEnumberF and -eturn: ex QCG
(3
Qx --Aet the +"-+ to x
Ax --Aet the ?D. to x
*x --Aet the s*ale to x
name --Aave the "lignment
=x --Aave 0oes bac& x it
return --A*ompute the "lignment
9 --Ae9it
E3+D-"+31. $F E+"-+5%CCF E?D.5 2GF E*"?D5-CGGF @1>- */13*D:
&f you want to indicate the coordinates, relative to a specific se3uence, type)
I'se,_name*:start
Type 5Pyour nameQ to save the current alignment, and e@tract a new motif.
Type L when you are done.
!utput Control
@eneric
!onventions 7egarding Filenames
stdout, stderr, stdin, no, $dev$null are valid filenames. They cause the corresponding
file to be output in stderr or stdout, for an input file, stdin causes the program to
re3uests the corresponding file through pipe. No causes a suppression of the output,
as does $dev$null.
%dentifying the +utput files automatically
&n the tIcoffee output, each output appears in a line)
,,,,, 73?D."#D BnameA +@PD B+ypeA 71-#"+ B7ormatA
-no_warning
1sage- no(#arning
$efault: /8itc)ed off
5uppresseswarning output.
Alignments
-outfile
1sage- outfile29out(aln file,default,no.
$efault:-outfile4default
&ndicates the name of the alignment output by tIcoffee. &f the default is used, the alignment
is named <your sequences>.aln
-output
1sage- output29for!at0,for!atB,$$$.
($
$efault:-output4clustal8
&ndicates the format used for outputting the -outfile.
5upported formats are)
clustalwIaln, clustalw ) Clustal' format.
gcg, msfIaln ) !5 alignment.
pirIaln ) pir alignment.
fastaIaln ) fasta alignment.
phylip ) (hylip format.
pirIse3 ) pir se3uences #no gap%.
fastaIse3 ) fasta se3uences #no gap%.
"s well as)
scoreIascii ) causes the output of a reliability flag
scoreIhtml ) causes the output to be a reliability plot in /T!L
scoreIpdf ) idem in (+ #if ps,pdf is installed on your system%.
scoreIps ) idem in postscript.
!ore than one format can be indicated)
PROMPT: t_coffee sample_se,A.fasta -output4clustal8KgcgK
score_)tml
" publication describing the CO2A inde@ is available on)
http)$$www.tcoffee.org$(ublications$(df$core.pp.pdf
-outse'weight
1sage- outse/#eight29filena!e.
$efault: not used
&ndicates the name of the file in which the se3uences weights should be saved..
-case
1sage- case29keep,upper,lo#er.
$efault: -case4Feep
&nstructs the program on the case to be used in the output file #Clustalw uses upper
case%. The default keeps the case and makes it possible to maintain a mi@ture of
upper and lower case residues.
&f you need to change the case of your file, you can use se3Ireformat)
((
PROMPT: t_coffee ot)er_pg se,_reformat in sample_alnA.aln
action Qlo8er output clustal8
-cpu
1sage- deprecated
-outse'weight
Hsage) -outse3weightMPname of the file containing the weights appliedQ
+efault) -outse3weightMno
'ill cause the program to output the weights associated with every se3uence in the
dataset.
-outorder [cw]
1sage- outorder29input &? aligned &? filena!e.
$efault:-outorder4input
5ets the order of the se3uences in the output alignment) outorder2input means the
se3uences are kept in the original order. outorder2aligned means the se3uences come in
the order indicated by the tree. This order can be seen as a one-dimensional proEection of the
tree distances. 5outdorder29filena!e.ilename is a legal fasta file, whose order will be
used in the final alignment.
-inorder [cw]
1sage- inorder29input &? aligned.
$efault:-inorder4aligned
!ultiple alignments based on dynamic programming depend slightly on the order in which
the incoming se3uences are provided. To prevent this effect se3uences are arbitrarily sorted
at the beginning of the program #-inorderMaligned%. /owever, this affects the se3uence order
within the library. Kou can switch this off by ststing ZinorderMinput.
-se'nos
1sage- se/nos29on or off.
$efault:-se,nos4off
Causes the output alignment to contain residue numbers at the end of each line)
+-*177DD
se!C aaa---aaaa--------aa I
se!% a-----aa-----------a 2
se!C a-----------------a CC
se!% aaaaaaaaaaaaaaaaaaa CI
4ibraries
"lthough, it does not necessarily do so e@plicitly, T-Coffee always end up
combining libraries. Libraries are collections of pairs of residues. 0iven a set of
libraries, T-Coffee makes an attempt to assemble the alignment with the highest
level of consistence. Kou can think of the alignment as a timetable. Aach library pair
would be a re3uest from students or teachers, and the Eob of T-Coffee would be to
(.
assemble the time table that makes as many people as possible happyO
-out_lib
Hsage) -outIlibMPname of the library,default,noQ
+efault)-outIlibMdefault
5ets the name of the library output. +efault implies PrunInameQ.tcIlib
-lib_only
1sage- lib(only
$efault: unset
Causes the program to stop once the library has been computed. !ust be used in conEunction
with the flag 5out(lib
Trees
-newtree
1sage- ne#tree29tree file.
$efault: 5o file specified
&ndicates the name of the file into which the guide tree will be written. The default will be
Pse3uenceInameQ.dnd, or PrunIname.dndQ. The tree is written in the parenthesis format
known as newick or New /ampshire and used by (hylips #see the format section%.
2o NOT con!se this (!ide tree %ith a ph#lo(enetic tree.
"eliability #stimation
C!"# Computation
The CO2A is an inde@ that indicates the consistency between the library of piarwise
alignments and the final multiple alignment. Our e@periment indicate that the higher
this consistency, the more reliable the alignment. " publication describing the
CO2A inde@ can be found on)
http)$$www.tcoffee.org$(ublications$(df$core.pp.pdf
-evaluate_mode
1sage-
e'aluate(!ode29t(coffee(fast,t(coffee(slo#,t(coffee(non(extende
d .
$efault: -ealuate_mode4t_coffee_fast
This flag indicates the mode used to normaliFe the tIcoffee score when computing the
reliability score.
t_co""ee_"ast) NormaliFation is made using the highest score in the !5". This evaluation
mode was validated and in our hands, pairs of residues with a score of : or higher have <- U
chances to be correctly aligned to one another.
t_co""ee_slow# NormaliFation is made using the library. This usually results in lower score
(/
and a scoring scheme more sensitive to the number of se3uences in the dataset. Note that this
scoring scheme is not any more slower, thanks to the implementation of a faster heuristic
algorithm.
t_co""ee_non_e&tended# the score of each residue is the ratio between the sum of its non
e@tended scores with the column and the sum of all its possible non e@tended scores.
These modes will be useful when generating colored version of the output, with the 5output
flag)
PROMPT: t_coffee sample_se,A.fasta ealuate_mode t_coffee_slo8
output score_asciiK score_)tml
PROMPT: t_coffee sample_se,A.fasta ealuate_mode t_coffee_fast
output score_asciiK score_)tml
PROMPT: t_coffee sample_se,A.fasta ealuate_mode
t_coffee_non_e"tended output score_asciiK score_)tml
@eneric !utput
-run_name
1sage- run(na!e29your run na!e.
$efault: no default set
This flag causes the prefi@ Pyour se3uencesQ to be replaced by Pyour run nameQ
when renaming the default output files.
-'uiet
1sage- /uiet29stderr,stdout,file na!e &? nothing.$
$efault:-,uiet4stderr
2edirects the standard output to either a file. /uiet on its own redirect the output to
$dev$null.
-align [!,]
This flag indicates that the program must produce the alignment. &t is here for
compatibility with Clustal'.
AP*%3 i"+* and t"+* Parameters
)arnin(: These 1a(s %ill onl# %or3 %ithin the AP24 pac3a(e that can be
in&o3ed &ia the +other_p( parameter o T-5o/ee:
t_co/ee +other_p( apdb +aln 6#o!r aln.
(6
-'uiet [6ame as 3-!offee]
-run_name [6ame as 3-!offee]
-aln
1sage- aln29file(na!e.$
$efault:none
&ndicates the name of the file containing the se3uences that need to be evaluated. The
se3uences whose structure is meant to be used must be named according to their (+4
identifier.
The format can be "5T", CLH5T"L or any of the formats supported by T-Coffee. "(+4
only evaluates residues in capital and ignores those in lower case. &f your se3uences are in
lower case, you can upper case them using se3Ireformat)
PROMPT: t_coffee ot)er_pg se,_reformat in 3d_sample;.aln action
Qupper output clustal8 * 3d_sample;.c8_aln
The alignment can then be evaluated using the defaultr of "(+4)
PROMPT: t_coffee ot)er_pg apd( aln 3d_sample;.aln
The alignment can contain as many structures as you wish.
-n_excluded_nb
1sage- n(excluded(nb29integer.$
$efault:A
'hen evaluating the local score of a pair of aligned residues, the residues immediately ne@t
to that column should not contribute to the measure. 4y default the first to the left and first
to the right are e@cluded.
-maximum_distance
1sage- !axi!u!(distance29float.$
$efault:A2
5iFe of the neighborhood considered around every residue. &f .-localImode is set to sphere,
-ma@imumIdistance is the radius of a sphere centered around each residue. &f ZlocalImode is
set to window, then Zma@imumIdistance is the siFe of the half window #i.e. windowIsiFeM-
ma@imumIdistanceJ,\9%.
-similarity_threshold
1sage- si!ilarity(threshold29integer.$
$efault:72
raction of the neighborhood that must be supportive for a pair of residue to be considered
correct in "(+4. The neighborhood is a sphere defined by Zma@imumIdistance, and the
support is defined by ZmdIthreshold.
(8
-local_mode
1sage- local(!ode29sphere,#indo#.$
$efault:sp)ere
+efines the shape of a neighborhood, either as a sphere or as a window.
-filter
1sage- filter29C$CC0$CC.$
$efault:A.22
+efines the centiles that should be kept when making the local measure. oir instance,
-filterM-.<- means that the the 9- last centiles will be removed from the evaluation. The
filtration is carried out on the i2!5+ values.
-print_rapdb [*nsupported]
1sage- print(rapdb 4)+AD6
$efault:off
This causes the prints out of the e@act neighborhood of every considered pair of residues.
-outfile [6ame as 3-!offee]
This flag is meant to control the output name of the colored "(+4 output. This file
will either display the local "(+4 score or the local i2!+, depending on the value
of ZcolorImode. The default format is defined by Zouptut and is scoreIhtml.
-color_mode
1sage- color(!ode29apdb, ir!sd.
$efault:apd(
This flag is meant to control the colored "(+4 output #local score%. This file will
either display the local "(+4 score or the local i2!+.
.:
%uilding a +er5er
'e maintain a T-Coffee server #www.tcoffee.org%. 'e will be pleased to provide
anyone who wants to set up a similar service with the sources
#n5ironment >ariables
T-Coffee stores a lots of information in locations that may be unsuitable when
running a server.
4y default, T-Coffee will generate and rely on the follwing directory structure)
/home/youraccount/ ,/1#D_2_+*177DD
/1#D_2_+*177DD/.t_coffee/ ,83-_2_+*177DD
83-_2_+*177DD/cache ,*"*/D_2_+*177DD
83-_2_+*177DD/tmp ,+#P_2_+*177DD
83-_2_+*177DD/methods ,#D+/1_2_+*177DD
83-_2_+*177DD/mcoffee ,#*177DD_2_+*177DD
4y default, all these directories are automatically created, following the
dependencies suggested here.
The first step is the determination of the /O!A. 4y default the program tries to use
/O!AI>ITCOAA, then the /O!A variable and T!( or TA!( if /O!A is not
set on your system or your account. &t is your responsibility to make sure that one of
these variables is set to some valid location where the T-Coffee process is allowed to
read and write.
&f no valid location can be found for /O!AI>ITCOAA, the program e@its. &f
you are running T-Coffee on a server, we recommend to hard set the following
locations, where your scratch is a valid location.
/1#D_2_+*177DD5Ryour scratchR
+#P_2_+*177DD5Ryour scratchR
83-_2_+*177DD5Ryour scratchR
*"*/D_2_+*177DD5Ryour scratchR
.1_D--1-_-DP1-+_2_+*177DD5C
Note that it is a good idea to have a cron Eob that cleans up this scratch area, once in
a while.
.1
!utput of the Bdnd fileB
" common source of error when running a server) T-Coffee !H5T output the .dnd
file because it re-reads it to carry out the progressive alignment. 4y default T-Coffee
outputs this file in the directory where the process is running. &f the T-Coffee
process does not have permission to write in that directory, the computation will
abort...
To avoid this, simply specify the name of the output tree)
-newtreeMPwritable file #usually in $tmp%Q
Chose the name so that two processes may not over-write each other dnd file.
Permissions
The tIcoffee process !H5T be allowed to write in some scratch area, even when it
is ran by !r nobody... !ake sure the $tmp$ partition is not protected.
!ther Programs
T-Coffee may call various programs while it runs #lalign,list by defaults%. !ake
sure your process knows where to find these e@ecutables.
.2
&ormats
Parameter files
(arameter files used with -parameters, -tIcoffeeIdefaults, -daliIdefaults... !ust
contain a valid parameter string where line breaks are allowed. These files cannot
contain any comment, the recommended format is one parameter per line)
Bparameter nameA5BvalueCA;Bvalue%A....
Bparameter nameA5.....
+e;uence 9ame ?andling
5e3uence name handling is meant to be fully consistent with Clustal' #Gersion
9.;:%. This implies that in some cases the names of your se3uences may be edited
when coming out of the program. ive rules apply)
Naming Your Sequences the Right Way
-'o (pace
'ames that do contain spaces) "or instance#
>seq human_myc
will *e turned into
>seq
+t is your responsi*ility to ma$e sure that the names you pro,ide are not am*iguous
a"ter such an editing. -his editing is consistent with .lustalw (/ersion .05)
2-'o (trange .haracter
(ome non alpha*etical characters are replaced with underscores. -hese are# 12#()1
3ther characters are legal and will *e $ept unchanged. -his editing is meant to $eep
in line with .lustalw (/ersion .05).
4-> is '5/56 legal (e&cept as a header to$en in a %7(-7 "ile)
8-'ame length must *e *elow 00 characters) although 5 is recommended "or
compati*ility with other programs.
5-9uplicated sequences will *e renamed (i.e. sequences with the same name in the
same dataset) are allowed *ut will *e renamed according to their original order.
:hen sequences come "rom multiple sources ,ia the ;in "lag) consistency o" the
.3
renaming is not guaranteed. <ou should a,oid duplicated sequences as they will cause
your input to di""er "rom your output thus ma$ing it di""icult to trac$ data.
Automatic &ormat "ecognition
!ost common formats are automatically recogniFed by tIcoffee. 5ee -in and the
ne@t section for more details. &f your format is not recogniFed, use readse3 or
clustalw to switch to another format. 'e recommend asta.
+tructures
(+4 format is recogniFed by T-Coffee. T-Coffee uses e@tractIfromIpdb #cf Z
otherIpg flag%. e@tractIfromIpdb is a small embeded module that can be used on its
own to e@tract information from pdb files.
"9A +tructures
2N" structures can either be coded as T-Coffee libraries, with each line indicating
two paired residues, or as alifold output. The sele@ format is also partly supported
#see the se3Ireformat tutorial on 2N" se3uences handling%.
+e;uences
5e3uences can come in the following formats) fasta, pir, swiss-prot, clustal aln, msf
aln and tIcoffee aln. These formats are the one automatically recogniFed. (lease
replace the DJD sign sometimes used for stop codons with an L.
Alignments
"lignments can come in the following formats) msf, Clustal', asta, (ir and
tIcoffee. The tIcoffee format is very similar to the Clustal' format, but slightly
more fle@ible. "ny interleaved format with se3uence name on each line will be
correctly parsed)
Bempy lineA E7acultativeFn
Bline of textA E-e!uiredF
Bline of textA E7acultativeFn
Bempty lineA E-e!uiredF
Bempty lineA E7acultativeFn
Bse!C nameABspaceABse!CA
Bse!% nameABspaceABse!%A
Bse!$ nameABspaceABse!$A
Bempty lineA E-e!uiredF
Bempty lineA E7acultativeFn
Bse!C nameABspaceABse!CA
Bse!% nameABspaceABse!%A
Bse!$ nameABspaceABse!$A
Bempty lineA E-e!uiredF
Bempty lineA E7acultativeFn
"n empty line is a line that does NOT contain amino-acid. " line that contains the
Clustal' annotation #.)J% is empty.
5paces are forbidden in the name. 'hen the alignment is being read, non character
signs are ignored in the se3uence field #such as numbers, annotationO%.
3ote- a different nu!ber of lines in the different blocks #ill cause the progra! to crash
.$
or hang$
4ibraries
T-C!&&##<4)%<&!"AT<:1
This is currently the only supported format.
SBspaceA +*_?3=_71-#"+_GC
Bnse!A
Bse!C nameA Bse!C lengthA Bse!CA
Bse!% nameA Bse!% lengthA Bse!%A
Bse!$ nameA Bse!$ lengthA Bse!$A
S*omment
:S*omment<n
,iC i%
-iC -i% 4C :4%; 4$<
,C %
C% C$ II :C%/G vs C$/C; weight II<
C% C2 PG
C) CL )L
,C $
C% C$ II
C% C2 PG
C) CL )L
SBspaceADN_C_+1_.
5i9) inde@ of 5e3uence 9
2i9) inde@ of residue 9 in se39
G9) &nteger Galue) 'eight
G,, G8) optional values
3ote 0- There is a space bet#een the ! And 7;L(0(T&(3
3ote B- The last line 4! 7;L(0(T&(36 indicates that-
5e3uences and residues are numbered from 9 to N, unless the token 5AVI9ITOIN
is omitted, in which case the se3uences are numbered from - to N-9, and residues
are from 9 to N.
2esidues do not need to be sorted, and neither do the se3uences. The same pair can
appear several times in the library. or instance, the following file would be legal)
,C %
C% C$ II
,C %
C) CL II
,C C
C% C2 PG
&t is also poosible to declare ranges of resdues rather than single pairs. or instance,
the following)
,G C
T=?1*MT CG C% C2 II
T=?1*MT C) $G 2G II
,G %
C) CL II
,G C
C% C2 PG
.(
The first statement 4LOCC declares a 4LOCC of length 9-, that starts on position
9, of se3uence 9 and position 9> of se3uence , and where each pair of residues
within the block has a score of <<. The second 4LOCC starts on residue 8- of 9,
residue >- of , and e@tends for 9: residues.
4locks can overalp and be incompatible with one another, Eust like single
constraints.
T-C!&&##<4)%<&!"AT<:2
" simpler format is being developed, however it is not yet fully supported and is
only mentioned here for development purpose.
S +*_?3=_71-#"+_G%
,C DNC E1P+31."?F
,% DN% E1P+31."?F
...
Scomment E1P+31."?F
C -C -iC % -% -i% 4C :4% 4$<
5A . -C -iC % -% -i% 4C :4% 4$<
...
59, 5,) name of se3uence 9 and ,
5AV9) se3uence of 59
2i9, 2i,) inde@ of the residues in their respective se3uence
29, 2,) 2esidue type
G9, G,, G8) integer Galues #G, and G8 are optional%
Galue9, Galue , and Galue8 are optional.
4ibrary 4ist
These are lists of pairs of se3uences that must be used to compute a library. The
format is)
Bnse!A BCA B%A
% hamg% globav
$ hamgw hemog singa
...
+ubstitution matricesB
&f the re3uired substitution matri@ is not available, write your own in a file using the
following format)
Clustal0 +tyle C*eprecatedD
, *?>+"?6_#"+-39 71-#"+
U
vC
v% v$
v2 v) vL
...
U
v9, v,... are integers, possibly negatives.
..
The order of the amino acids is) "4C+A0/&CL!NV25TG'LK^, which means
that v9 is the substitution value for " vs ", v, for " vs 4, v8 for 4 vs 4, v> for " vs
C and so on.
%4A+T &ormat C"ecommendedD
, =?"+_#"+-39 71-#"+
, "?P/"=D+5"0*+
" 0 * +
" G C % $
0 G % $ 2
* C C % $
...
The alphabet can be freely defined
+e;uences 0eights
Create your own weight file, using the -se3Iweight flag)
, 3.0?D_DN_6D30/+_71-#"+_GC
se!_nameC vC
se!_name% v%
...
No duplicate allowed. 5e3uences not included in the set of se3uences provided to
tIcoffee will be ignored. Order is free. G9 is a float. Hn-weighted se3uences will see
their weight set to 9.
./
Eno1n Problems
9-5ensitivity to se3uence order) &t is difficult to implement a !5" algorithm totally
insensitive to the order of input of the se3uences. &n tIcoffee, robustness is
increased by sorting the se3uences alphabetically before aligning them. 4eware that
this can result in confusing output where se3uences with similar name are
une@pectedly close to one another in the final alignment.
,-Nucleotides se3uences with long stretches of Ns will cause problems to lalign,
especially when using !occa. To avoid any problem, filter out these nucleotides
before running mocca.
8-5top codons are sometimes coded with DJD in protein se3uences. This will cause
the program to crash or hang. (lease replace the DJD signs with an L.
>-2esults can differ from one architecture to another, due rounding differences. This
is caused by the tree estimation procedcure. &f you want to make sure an alignment
is reproducible, you should keep the associated dendrogram.
:-+eploying the program on a
.6
Technical 9otes
These notes are only meant for internal development.
*e5elopment
The following e@amples are only meant for internal development, and are used to
insure stability from release to release
(2O& LA,L& 5T
prf9) profile containing one structure
prf,) profile containing one structure
PROMPT: t_coffee Rsample_profileA.alnKRsample_profileD.aln
-mode43dcoffee -outfile4aligned_prf.aln
Command 4ine 4ist
These command lines have been checked before every release #along with the other
CL in this documentation)
-e@ternal methods7
PROMPT: t_coffee sample_se,A.fasta
in4Mclustal8_pairKMclustal8_msaKMslo8_pair outfile4clustal_te"t
-fugueIclient
PROMPT: t_coffee in /sample_se,B.fasta Pstruc;.pd( Mfugue_pair
-" list of command lines kindly provided by 1ames 'atson #used to crash the pg
before version 8.>-%
.8
PROMPT: t_coffee -in /se,.fas PDPT6 Mfugue_pair
PROMPT: t_coffee -in /Dse,s.fas Mfugue_pair -template_file /E1<_P_
PROMPT: t_coffee -mode 3dcoffee -in /se,.fas PDPT6
PROMPT: t_coffee -mode 3dcoffee -in /Dse,s.fas -template_file
/E1<_P_
-" list of command lines that crashed the program before 8.69
PROMPT: t_coffee sample_se,3.fasta in Mfast_pair Msap_pair
Mfugue_pair template_file template_file3.template
-A commad lie !o "ead B"ela;ed> pd$ @les,,,
PROMPT: t_coffee in Msap_pair /sample_se,7.fasta template_file
template_file7.template 8eig)t A22A out_li( test_li(7.tc_li(
li(_onl+
--a"si% o9 *ARNA li$"a"ies
PROMPT: t_coffee in 1marna.tc_li( outfile maran.test
--a"si% o9 lo% se6#ece lies(
PROMPT: t_coffee in 0sample_alnB.aln outfile test.aln
/:
To *o
-implement H(0!" tree computation
-implement se3,dpaItree
-debug dpa
-2econciliate se3uences and template when reading the template
-"dd the server command lines to the checking procedure
/1
Das könnte Ihnen auch gefallen
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceVon EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceBewertung: 4 von 5 Sternen4/5 (895)
- Never Split the Difference: Negotiating As If Your Life Depended On ItVon EverandNever Split the Difference: Negotiating As If Your Life Depended On ItBewertung: 4.5 von 5 Sternen4.5/5 (838)
- The Yellow House: A Memoir (2019 National Book Award Winner)Von EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Bewertung: 4 von 5 Sternen4/5 (98)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeVon EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeBewertung: 4 von 5 Sternen4/5 (5794)
- Shoe Dog: A Memoir by the Creator of NikeVon EverandShoe Dog: A Memoir by the Creator of NikeBewertung: 4.5 von 5 Sternen4.5/5 (537)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaVon EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaBewertung: 4.5 von 5 Sternen4.5/5 (266)
- The Little Book of Hygge: Danish Secrets to Happy LivingVon EverandThe Little Book of Hygge: Danish Secrets to Happy LivingBewertung: 3.5 von 5 Sternen3.5/5 (400)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureVon EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureBewertung: 4.5 von 5 Sternen4.5/5 (474)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryVon EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryBewertung: 3.5 von 5 Sternen3.5/5 (231)
- Grit: The Power of Passion and PerseveranceVon EverandGrit: The Power of Passion and PerseveranceBewertung: 4 von 5 Sternen4/5 (588)
- The Emperor of All Maladies: A Biography of CancerVon EverandThe Emperor of All Maladies: A Biography of CancerBewertung: 4.5 von 5 Sternen4.5/5 (271)
- The Unwinding: An Inner History of the New AmericaVon EverandThe Unwinding: An Inner History of the New AmericaBewertung: 4 von 5 Sternen4/5 (45)
- On Fire: The (Burning) Case for a Green New DealVon EverandOn Fire: The (Burning) Case for a Green New DealBewertung: 4 von 5 Sternen4/5 (74)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersVon EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersBewertung: 4.5 von 5 Sternen4.5/5 (345)
- Team of Rivals: The Political Genius of Abraham LincolnVon EverandTeam of Rivals: The Political Genius of Abraham LincolnBewertung: 4.5 von 5 Sternen4.5/5 (234)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreVon EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreBewertung: 4 von 5 Sternen4/5 (1090)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyVon EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyBewertung: 3.5 von 5 Sternen3.5/5 (2259)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)Von EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Bewertung: 4.5 von 5 Sternen4.5/5 (121)
- Her Body and Other Parties: StoriesVon EverandHer Body and Other Parties: StoriesBewertung: 4 von 5 Sternen4/5 (821)
- HFM Installation DacumentDokument50 SeitenHFM Installation DacumentR SreenuNoch keine Bewertungen
- PF600 User GuideDokument56 SeitenPF600 User Guide徐木松Noch keine Bewertungen
- Train Borne ATP GuideDokument12 SeitenTrain Borne ATP Guidevineet_k_signalNoch keine Bewertungen
- IndiaFoxtEcho Eurofighter Typhoon ManualDokument108 SeitenIndiaFoxtEcho Eurofighter Typhoon ManuallaniusNoch keine Bewertungen
- Install The Required Software: RequirementsDokument5 SeitenInstall The Required Software: Requirementsallen edvNoch keine Bewertungen
- Installation - NQWebDokument9 SeitenInstallation - NQWebMete TorunNoch keine Bewertungen
- TC300-350 User ManualDokument125 SeitenTC300-350 User ManualRoberto MunozNoch keine Bewertungen
- Logix Um003 - en PDokument113 SeitenLogix Um003 - en PVinícius LemosNoch keine Bewertungen
- MyQ 5-2 Install Manual EN Rev46Dokument163 SeitenMyQ 5-2 Install Manual EN Rev46Gaston LinNoch keine Bewertungen
- 2user Manual ARCTIC USB Dongle UD2 r1 enDokument2 Seiten2user Manual ARCTIC USB Dongle UD2 r1 enLex LexisNoch keine Bewertungen
- Paragon ManualDokument22 SeitenParagon ManualMarcelo Vourakis BarbosaNoch keine Bewertungen
- iCR User Manual v9.7Dokument254 SeiteniCR User Manual v9.7Slingbox VCMediosNoch keine Bewertungen
- Smartplant Electrical: Release BulletinDokument24 SeitenSmartplant Electrical: Release Bulletinvenkateee57Noch keine Bewertungen
- Installation of Microwin Programming SoftwareDokument18 SeitenInstallation of Microwin Programming SoftwareAndrewNoch keine Bewertungen
- NotePerformer - Users GuideDokument51 SeitenNotePerformer - Users GuideRodrigo Morales MaldonadoNoch keine Bewertungen
- YDA0004N1 How To Install CAMIO 8.0 Software - EngDokument34 SeitenYDA0004N1 How To Install CAMIO 8.0 Software - EngMememetNoch keine Bewertungen
- Installing Destiny: Installation RequirementsDokument9 SeitenInstalling Destiny: Installation RequirementsarrondeshazoNoch keine Bewertungen
- Civil 3 D2016 Pro Pack 3 READMEDokument3 SeitenCivil 3 D2016 Pro Pack 3 READMEOscar RodríguezNoch keine Bewertungen
- Instruction NETLink-lite 1.04 PDFDokument37 SeitenInstruction NETLink-lite 1.04 PDFstudskirbyNoch keine Bewertungen
- Burrtools-0 6 3-A4Dokument91 SeitenBurrtools-0 6 3-A4javimmNoch keine Bewertungen
- USB Cable of MSR206 Series Magnetic Card Reader Writer Installation GuideDokument14 SeitenUSB Cable of MSR206 Series Magnetic Card Reader Writer Installation GuidebootybethathangNoch keine Bewertungen
- GHS TrainingBookDokument84 SeitenGHS TrainingBookKelvin XuNoch keine Bewertungen
- Techlog - Installation and Licensing Guide PDFDokument22 SeitenTechlog - Installation and Licensing Guide PDFBOUMERAOUNoch keine Bewertungen
- Autofx Photographic Edges 6.0 ManualDokument111 SeitenAutofx Photographic Edges 6.0 ManualFelze100% (1)
- Flares User GuideDokument43 SeitenFlares User GuidesddrlNoch keine Bewertungen
- GEOTRAN FWD AnalysisDokument19 SeitenGEOTRAN FWD Analysisdigvijay singhNoch keine Bewertungen
- Lpile 6 User Manual, Version 1Dokument97 SeitenLpile 6 User Manual, Version 1Daniyal AhmadNoch keine Bewertungen
- 2.1 Installation Requirements - . - . - . - . - . - . - . 3Dokument42 Seiten2.1 Installation Requirements - . - . - . - . - . - . - . 3Hugo Fernando SolisNoch keine Bewertungen
- Arduino Debug ToolDokument22 SeitenArduino Debug ToolSofyan HalwaNoch keine Bewertungen
- Solve Me If You CanDokument6 SeitenSolve Me If You CanSujithNoch keine Bewertungen