Beruflich Dokumente
Kultur Dokumente
Final Version - Surf 2016 Rat-Crispr-Prh Poster Se
Hochgeladen von
api-299660206Originaltitel
Copyright
Verfügbare Formate
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenCopyright:
Verfügbare Formate
Final Version - Surf 2016 Rat-Crispr-Prh Poster Se
Hochgeladen von
api-299660206Copyright:
Verfügbare Formate
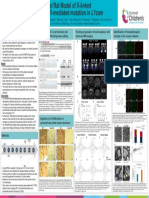
CRISPR/Cas9-Based Development of Novel Transgenic Rat Model of
Congenital Hydrocephalus
A. Scott Emmert1, Crystal Shula1, Francesco Mangano1, Kenneth Campbell1,2, Rolf Stottmann1,3, Yinhuai Chen4, Yueh-Chiang Hu4, and June Goto1
Div. of Pediatric Neurosurgery1, Div. of Developmental Biology2, Div. of Human Genetics3, and Transgenic Animal and Genome Editing Core Facility4, CCHMC
prh
PX459
Results
A
(-) strand sequence
(-) strand sequence
g37
EGP
Figure 1. Electroporation of C6 rat glial cells using the Neon Transfection System.
(+) strand sequence
Figure 5. Design of rat-CRISPR-prh gRNA, primers, and oligonucleotide donor template for introduction of the same single nucleotide
change found in prh mice into rat embryo.
A guide RNA (gRNA) sequence and oligonucleotide donor template
specific to the Ccdc39 gene mutation were designed (Optimized
CRISPR Design) to target a single nucleotide change in the rat
genome. Forward and reverse primers were similarly designed for
insertion of gRNA into a pSpCas9(BB)-2A-Puro (PX459) plasmid
construct.
PX459 plasmid was plated with DH5 competent cells onto LB/amp
agar plates and incubated at 37 for 16 hours. PX459 DNA was
then purified from a transformed bacterial colony and
spectrophotometrically analyzed.
BBSI restriction endonuclease was used to digest PX459, which
was then analyzed for the presence of cleaved bands using gel
electrophoresis. Digested PX459 purified from gel was treated with
alkaline phosphatase.
Diluted insert containing Rat-prh-F and Rat-prh-R primers was
ligated into the PX459 vector to create the PX459-rat-CRISPR-prh
plasmid. DH5 competent cells were transformed with PX459-ratCRISPR-prh, plated, and incubated.
PX459-rat-CRISPR-prh plasmid DNA was purified from the
transformed bacterial culture and analyzed spectrophotometrically.
PX459-rat-CRISPR-prh plasmid was sequenced for correct
insertion and ligation of the gRNA target sequence.
Colonies with positive sequencing results were cultured. PX459-ratCRISPR-prh (rat-prh), PX459, px458-g37 (g37, CRISPR plasmid
positive control), and pEGFP-N1(EGP) DNA was then purified.
Using the Neon Transfection System, C6 cells derived from rat glial
tumor were transfected with rat-prh, PX459, g37, and EGP (Figure
1). EGP fluorescence was used to measure transfection efficiency.
Genomic DNA (gDNA) was extracted from the transfected cells
using the Maxwell 16 System.
Rat-prh and g37 gDNA was amplified with rat-prh assay primers
and g37 assay primers through PCR, and the SURVEYOR
mismatch cleavage assay was used to assess the editing activity of
the rat-CRISPR-prh gRNA construct compared to that of the g37
gRNA positive control (Figure 2).
prh-1
prh-2
g37-2
prh-1
prh-2
g37-1
g37-2
C+G
g37-1
prh-primer
g37-primer
G only
g37-2
prh-primer
prh-1
prh-2
g37-1
g37-2
C+G
Clustered Regularly Interspaced Short
Palindromic Repeats (CRISPR)
Bacterial immune system modified for
genome engineering
Three components:
1. gRNA
Scaffold: Cas9 binding
Spacer: Genomic target
2. RNA-guided Cas9 endonuclease
3. DNA repair template containing desired
sequence
Cas9-gRNA complex binds genomic target
sequence immediately upstream PAM (5
NGG 3 for S. pyogenes Cas9), Cas9
cleaves target with double stranded break
(DSB), and homology directed repair
Image adapted from Addgene: CRISPR/Cas9 Guide
https://www.addgene.org/CRISPR/guide/
(HDR) generates specific nucleotide
Image adapted from Addgene: CRISPR/Cas9 Guide,
https://www.addgene.org/CRISPR/guide/
change.
Figure 3. CRISPR/Cas9 targeted genome editing using homology directed repair
(HDR).
Progressive hydrocephaly, prh
mouse mutant displaying severe
hydrocephalus at 14-postnatal days
(P14) as compared to wild type
following ENU screen (Stottmann et
al., 2011).
The prh mouse mutants are born with
mild ventriculomegaly but develop
massive hydrocephalus and white
matter degeneration (indicated by
asterisks, F) by P14.
Recently, the possible causal
mutation was found within Ccdc39
Ref. Stottmann et al., Focusing Forward Genetics: A Tripartite ENU Screen for
Neurodevelopmental Mutations in the Mouse Genetics (2011)
gene (unpublished).
Figure 4. Progressive hydrocephaly, prh mouse mutant line isolated in a
forward genetic screen using ENU mutagenesis.
G only
Methods
Background
C only
g37-primer
C only
Figure 2. Process by which SURVEYOR mismatch cleavage assay validates
genetic modification delivered by Cas9-gRNA complex.
prh-1
Image adapted from Surveyor Nuclease Assay, https://en.wikipedia.org/wiki/Surveyor_nuclease_assay
A. Using Optimized CRISPR Design (Zhang Lab, MIT 2015), a guide RNA was designed to target the critical thymine nucleotide required for proper splicing
of the rat Ccdc39 gene (chr2: 139960533A, rn5), which is mutated in the prh mouse model of congenital hydrocephalus. The gRNA sequence must lie
immediately upstream the PAM sequence (5 TGG 3) for Cas9 to bind target DNA. Guide #4 was selected as the most efficient gRNA design for its high ontarget quality score and low off-target activity score. Rat-CRISPR-prh forward and reverse primers were similarly designed to include the Guide #4 gRNA
targeting sequence for insertion using the PX459 vector. B. Negative (PAM-containing) and positive (non-PAM-containing) strand sequences of donor
oligonucleotide repair template were constructed with substitution of thymine to adenine in target sequence. Donor oligonucleotide was designed with
HindIII restriction site as SNP genotyping strategy.
prh-2
Congenital hydrocephalus is a devastating birth defect defined by
the abnormal accumulation of cerebrospinal fluid (CSF) in the brain.
Buildup of CSF places enormous pressure on the infants brain,
ultimately leading to brain damage and mental and physical
problems if left untreated. Although surgical intervention remains the
primary form of treatment for congenital hydrocephalus, it is not a
cure for this condition.
Emergence of the novel CRISPR/Cas9 genome-editing platform
provides an accessible and efficient method for investigating
possible gene mutations responsible for congenital hydrocephalus.
Through whole-genome sequencing, our group recently identified a
splice mutation within Ccdc39 (Coiled-coil domain containing 39)
gene as a strong mutation candidate for the mutant phenotype in a
congenital hydrocephalus mouse model, progressive hydrocephaly,
prh.
Defects in Ccdc39 gene disrupt normal motility of cilia and are the
cause of primary ciliary dyskinesia in humans and dogs.
Using the CRISPR/Cas9 system of targeted genome editing, this
study aims to introduce the same mutation evidenced in the prh
mouse mutant line into the rat genome.
Creation of a prh rat model of congenital hydrocephalus provides a
method to evaluate whether the mutation in Ccdc39 is responsible
for the hydrocephalus phenotype. Also, the prh rat model presents
opportunities to explore new surgical and imaging techniques and
to analyze the molecular functions of the Ccdc39 gene in rodent
brain development.
Methods (cont.)
g37-1
Introduction
900
800
700
900
800
700
600
500
400
600
500
300
400
200
300
100
200
100
2% gel 30 min
2% gel 1 hr
Figure 6. SURVEYOR mismatch cleavage assay confirms ability of PX459-rat-CRISPR-prh plasmid construct to modify genome of C6
rat glial cells following CRISPR/Cas9 genome editing.
Gel electrophoresis analysis of rat-CRISPR-prh and g37 digested DNA products using SURVEYOR mismatch cleavage assay. Prh and g37 DNA amplified
with corresponding g37-T7E1 and prh-T7E1 assay primers display cleaved bands indicative of SNP or indel in region of interest. Prh/prh-primer PCR
product at 888 bp is cleaved into 569 bp and 319 bp bands (red ovals) as initially designed into rat-CRISPR-prh T7E1 assay primers, demonstrating the
introduction of the single nucleotide change from thymine to adenine into rat genome as originally found in prh mice.
Conclusions
PX459-rat-CRISPR-prh plasmid and gRNA construct, which were designed to introduce the point mutation from thymine to adenine found in
prh mice into rat embryos, are capable of modifying the rat genome.
The successful result of this gRNA editing activity test in C6 cells demonstrates that the CRISPR/Cas9 platform can be used to generate a prh
mutant rat model of congenital hydrocephalus. This platform for targeted genome editing provides an efficient and adaptable method for
studying the molecular mechanisms of genetic diseases.
gRNA design programs like Optimized CRISPR Design can be used to create CRISPR/Cas9 experiments that readily and precisely target
nearly any sequence in the genome.
Future Directions
Prh mutant rat line is currently being generated with help from the Transgenic Animal and Genome Editing Core Facility. Zygote injections of
rat-CRISPR-prh plasmid construct, gRNA, and donor oligonucleotide into Sprague Dawley rats are underway.
Rat mutants displaying progressive hydrocephaly, prh phenotype will be genotyped, surgically shunted by neurosurgeons from the Division
of Pediatric Neurosurgery, and analyzed using diffusion tensor imaging (DTI).
Evidence of hydrocephalic prh rats following introduction of the same single nucleotide change found in prh mice would strengthen the
hypothesis of this point mutation as the mutation candidate of the Ccdc39 splice mutation.
Das könnte Ihnen auch gefallen
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeVon EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeBewertung: 4 von 5 Sternen4/5 (5794)
- Ase 56x36 Surf 2018 Capstone PosterDokument1 SeiteAse 56x36 Surf 2018 Capstone Posterapi-299660206Noch keine Bewertungen
- Shoe Dog: A Memoir by the Creator of NikeVon EverandShoe Dog: A Memoir by the Creator of NikeBewertung: 4.5 von 5 Sternen4.5/5 (537)
- SRHSB L1cam Crispr Poster Post-Lab Meeting Final Version 35 X 47-MinDokument1 SeiteSRHSB L1cam Crispr Poster Post-Lab Meeting Final Version 35 X 47-Minapi-299660206Noch keine Bewertungen
- Pediatric Hernia Report Int 3041Dokument3 SeitenPediatric Hernia Report Int 3041api-299660206Noch keine Bewertungen
- The Yellow House: A Memoir (2019 National Book Award Winner)Von EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Bewertung: 4 von 5 Sternen4/5 (98)
- Scottie Emmert Honors Self-Designed Proposal Biomedical RampDokument6 SeitenScottie Emmert Honors Self-Designed Proposal Biomedical Rampapi-299660206Noch keine Bewertungen
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceVon EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceBewertung: 4 von 5 Sternen4/5 (895)
- Particle Bombartment PPT Tomorw - PPT - FinishingDokument52 SeitenParticle Bombartment PPT Tomorw - PPT - FinishingUma Rajagopalan100% (1)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersVon EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersBewertung: 4.5 von 5 Sternen4.5/5 (344)
- Analgesia Epidural Current Views and ApproachesDokument168 SeitenAnalgesia Epidural Current Views and ApproachesManuel Ortega100% (2)
- The Little Book of Hygge: Danish Secrets to Happy LivingVon EverandThe Little Book of Hygge: Danish Secrets to Happy LivingBewertung: 3.5 von 5 Sternen3.5/5 (399)
- Chidambaram Curriculum VitaeDokument3 SeitenChidambaram Curriculum VitaeasdfghjklNoch keine Bewertungen
- Grit: The Power of Passion and PerseveranceVon EverandGrit: The Power of Passion and PerseveranceBewertung: 4 von 5 Sternen4/5 (588)
- Basic Newborn Assessment FlashcardsDokument7 SeitenBasic Newborn Assessment FlashcardsChristina Chaston MonteithNoch keine Bewertungen
- The Emperor of All Maladies: A Biography of CancerVon EverandThe Emperor of All Maladies: A Biography of CancerBewertung: 4.5 von 5 Sternen4.5/5 (271)
- Turkey Your Partner in Health CareDokument22 SeitenTurkey Your Partner in Health Caremah927Noch keine Bewertungen
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaVon EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaBewertung: 4.5 von 5 Sternen4.5/5 (266)
- Advanced Practice Nursing in The United StatesDokument6 SeitenAdvanced Practice Nursing in The United StatesrhinoNoch keine Bewertungen
- Never Split the Difference: Negotiating As If Your Life Depended On ItVon EverandNever Split the Difference: Negotiating As If Your Life Depended On ItBewertung: 4.5 von 5 Sternen4.5/5 (838)
- تقويم الاسنان مقدمة 01Dokument6 Seitenتقويم الاسنان مقدمة 01Ibrahim TamlawiNoch keine Bewertungen
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryVon EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryBewertung: 3.5 von 5 Sternen3.5/5 (231)
- ZgmedDokument15 SeitenZgmedAkin AkinmosinNoch keine Bewertungen
- Angiofibroma NasofaringDokument4 SeitenAngiofibroma NasofaringRick RomeroNoch keine Bewertungen
- On Fire: The (Burning) Case for a Green New DealVon EverandOn Fire: The (Burning) Case for a Green New DealBewertung: 4 von 5 Sternen4/5 (73)
- Centennial LecturesDokument2 SeitenCentennial LectureshrhdanieNoch keine Bewertungen
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureVon EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureBewertung: 4.5 von 5 Sternen4.5/5 (474)
- Pediatrician Power Point 1Dokument11 SeitenPediatrician Power Point 1Ashley100% (1)
- Team of Rivals: The Political Genius of Abraham LincolnVon EverandTeam of Rivals: The Political Genius of Abraham LincolnBewertung: 4.5 von 5 Sternen4.5/5 (234)
- OK WRTK Resource DirectoryDokument101 SeitenOK WRTK Resource DirectoryTeddy WilsonNoch keine Bewertungen
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyVon EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyBewertung: 3.5 von 5 Sternen3.5/5 (2259)
- The Contribution of IMGs To The NHS PDFDokument47 SeitenThe Contribution of IMGs To The NHS PDFCrhistian Toribio DionicioNoch keine Bewertungen
- StemPro® Adipogenesis Differentiation KitDokument2 SeitenStemPro® Adipogenesis Differentiation KitTabita Timeea ScutaruNoch keine Bewertungen
- Shoulder DystociaDokument13 SeitenShoulder Dystociarolla_hiraNoch keine Bewertungen
- Spine 2019 BrochureDokument10 SeitenSpine 2019 BrochureLuciana AliceNoch keine Bewertungen
- 16 Prolapse SomaliDokument5 Seiten16 Prolapse SomaliMohamed AliNoch keine Bewertungen
- The Unwinding: An Inner History of the New AmericaVon EverandThe Unwinding: An Inner History of the New AmericaBewertung: 4 von 5 Sternen4/5 (45)
- Enhancing UK Core Medical Training Through Simulation-Based Education: An Evidence-Based ApproachDokument36 SeitenEnhancing UK Core Medical Training Through Simulation-Based Education: An Evidence-Based ApproachAndxp51Noch keine Bewertungen
- Kit GeriaDokument59 SeitenKit GeriaNila HasrializaNoch keine Bewertungen
- Resume2017 FinalDokument2 SeitenResume2017 Finalapi-347484976Noch keine Bewertungen
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreVon EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreBewertung: 4 von 5 Sternen4/5 (1090)
- The Early History of X-Ray Diagnosis - Mould PDFDokument47 SeitenThe Early History of X-Ray Diagnosis - Mould PDFClaudio MagalhaesNoch keine Bewertungen
- Notification Alteration of Two WheelersDokument8 SeitenNotification Alteration of Two WheelersSandip LulekarNoch keine Bewertungen
- Cleveland Clinic Abu Dhabi Residency Training ProgramDokument8 SeitenCleveland Clinic Abu Dhabi Residency Training ProgrampaulaNoch keine Bewertungen
- The Local Health Referral System ManualDokument72 SeitenThe Local Health Referral System ManualRuth Hazel Galang100% (1)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)Von EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Bewertung: 4.5 von 5 Sternen4.5/5 (120)
- Madison Solomon ResumeDokument2 SeitenMadison Solomon Resumeapi-321571692Noch keine Bewertungen
- International Institute of Reproduction and Fertility TrainingDokument6 SeitenInternational Institute of Reproduction and Fertility Trainingapi-294745900Noch keine Bewertungen
- Guidelines On: Paediatric UrologyDokument72 SeitenGuidelines On: Paediatric UrologyPatrascu CristiNoch keine Bewertungen
- 65 International Dr. Greenville, SC 29615Dokument4 Seiten65 International Dr. Greenville, SC 29615api-394232787Noch keine Bewertungen
- Mak 187 BulDokument4 SeitenMak 187 BulxephilimNoch keine Bewertungen
- Healthcare Practice Office Manager in New York NY Resume Milean CasianoDokument2 SeitenHealthcare Practice Office Manager in New York NY Resume Milean CasianoMileanCasiano100% (1)
- Her Body and Other Parties: StoriesVon EverandHer Body and Other Parties: StoriesBewertung: 4 von 5 Sternen4/5 (821)