Beruflich Dokumente
Kultur Dokumente
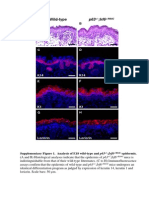
7.3. FASTA Format - 2
Hochgeladen von
csodaelme0 Bewertungen0% fanden dieses Dokument nützlich (0 Abstimmungen)
23 Ansichten2 SeitenFASTA format is a simple text-based format used to store and exchange DNA and protein sequences between programs. It consists of a single-line description beginning with ">" followed by lines of sequence data. Sequences are represented using standard IUB/IUPAC amino acid and nucleic acid codes with lowercase letters equivalent to uppercase. Dashes and asterisks may represent gaps and ambiguous characters respectively in some programs that recognize FASTA format.
Originalbeschreibung:
fasta
Originaltitel
7.3. FASTA Format_2
Copyright
© © All Rights Reserved
Verfügbare Formate
PDF, TXT oder online auf Scribd lesen
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenFASTA format is a simple text-based format used to store and exchange DNA and protein sequences between programs. It consists of a single-line description beginning with ">" followed by lines of sequence data. Sequences are represented using standard IUB/IUPAC amino acid and nucleic acid codes with lowercase letters equivalent to uppercase. Dashes and asterisks may represent gaps and ambiguous characters respectively in some programs that recognize FASTA format.
Copyright:
© All Rights Reserved
Verfügbare Formate
Als PDF, TXT herunterladen oder online auf Scribd lesen
0 Bewertungen0% fanden dieses Dokument nützlich (0 Abstimmungen)
23 Ansichten2 Seiten7.3. FASTA Format - 2
Hochgeladen von
csodaelmeFASTA format is a simple text-based format used to store and exchange DNA and protein sequences between programs. It consists of a single-line description beginning with ">" followed by lines of sequence data. Sequences are represented using standard IUB/IUPAC amino acid and nucleic acid codes with lowercase letters equivalent to uppercase. Dashes and asterisks may represent gaps and ambiguous characters respectively in some programs that recognize FASTA format.
Copyright:
© All Rights Reserved
Verfügbare Formate
Als PDF, TXT herunterladen oder online auf Scribd lesen
Sie sind auf Seite 1von 2
FASTA Format
FASTA format is a compact and simple method of storing DNA
and protein sequences as text files that can be read by virtually all
molecular biology programs.
A sequence in FASTA format
begins with a single-line description (or header),
followed by lines of sequence data.
The description line is distinguished from the sequence data
by a greater-than (">") symbol in the first column. It is
recommended that all lines of text be shorter than 80
characters in length.
An example sequence in FASTA format is:
essential!!!
name of the sequence
>gi|532319|pir|TVFV2E|TVFV2E envelope protein
ELRLRYCAPAGFALLKCNDADYDGFKTNCSNVSVVHCTNLMNTTVTTGLLLNGSYSENRT
QIWQKHRTSNDSALILLNKHYNLTVTCKRPGNKTVLPVTIMAGLVFHSQKYNLRLRQAWC
HFPSNWKGAWKEVKEEIVNLPKERYRGTNDPKRIFFQRQWGDPETANLWFNCHGEFFYCK
MDWFLNYLNNLTVDADHNECKNTSGTKSGNKRAPGPCVQRTYVACHIRSVIIWLETISKK
TYAPPREGHLECTSTVTGMTVELNYIPKNRTNVTLSPQIESIWAAELDRYKLVEITPIGF
APTEVRRYTGGHERQKRVPFVVQSQHLLAGILQQQKNLLAAVEAQQQMLKLTIWGVK
Sequences are expected to be represented in the standard IUB/IUPAC amino
acid and nucleic acid codes,
A Ala L Leu
B Asx M Met
C Cys N Asn
D Asp P Pro
E Glu Q Gln
F Phe R Arg
G Gly S Ser
H His T Thr
I Ile V Val
K Lys W Trp
Y Tyr
lower-case letters are are equivalent to upper-case.
Some, but not all programs that accept 'FASTA Format" recognize
- a hyphen or dash (-) to represent a gap of indeterminate length and
- an asterix (*) to represent an unknown or ambiguous character.
Introduction to Bioinformatics
Matthias Sipiczki, Department of Genetics, University of Debrecen
Comments to: lipovy@tigris.unideb.hu
Das könnte Ihnen auch gefallen
- Aat4422 Data s1Dokument12 SeitenAat4422 Data s1ar mcNoch keine Bewertungen
- ParpDokument1 SeiteParpapeachNoch keine Bewertungen
- Natural Selection at The RASGEF1C (GGC) Repeat in Human and Divergent Genotypes in Late Onset Neurocognitive DisorderDokument8 SeitenNatural Selection at The RASGEF1C (GGC) Repeat in Human and Divergent Genotypes in Late Onset Neurocognitive DisorderAnahí TessaNoch keine Bewertungen
- Pags CerdasDokument8 SeitenPags CerdasANGIE LORENA PALMA NI�ONoch keine Bewertungen
- What Do We Realize Past Trustworthiness in VoxelBased Enrollment Validation in The Exactness Regarding Regional VoxelBased Registration RVBR Methods For Orthognathic Surgical Procedure AnalysiscluoqDokument2 SeitenWhat Do We Realize Past Trustworthiness in VoxelBased Enrollment Validation in The Exactness Regarding Regional VoxelBased Registration RVBR Methods For Orthognathic Surgical Procedure Analysiscluoqmaleroute12Noch keine Bewertungen
- Aman NyeDokument13 SeitenAman NyeSolah AlaamNoch keine Bewertungen
- Sensitive SYBR Green-Real Time PCR For The Detection and Quantitation of Avian Rotavirus ADokument9 SeitenSensitive SYBR Green-Real Time PCR For The Detection and Quantitation of Avian Rotavirus ABertha Catalina RostroNoch keine Bewertungen
- Second Messenger Generation and Destruction: OCTOBER 1992Dokument5 SeitenSecond Messenger Generation and Destruction: OCTOBER 1992sant_grNoch keine Bewertungen
- 9100 BTS DescriptionDokument5 Seiten9100 BTS Description123 mlmbNoch keine Bewertungen
- A Semi Synthetic Organism II Carolina Nature 2017Dokument20 SeitenA Semi Synthetic Organism II Carolina Nature 2017José Antonio Silva NetoNoch keine Bewertungen
- Practicals in GeneticsDokument112 SeitenPracticals in GeneticscarlosaeserranoNoch keine Bewertungen
- Pi Is 0021925819692823Dokument13 SeitenPi Is 0021925819692823Robert BiliboacaNoch keine Bewertungen
- Pan Et Al 2019Dokument8 SeitenPan Et Al 2019misilpiyapiyoNoch keine Bewertungen
- Paraparesia Espástica - Estudo BrasilDokument3 SeitenParaparesia Espástica - Estudo BrasilLaura A M MNoch keine Bewertungen
- NeomycinDokument3 SeitenNeomycinDilek DabanlıNoch keine Bewertungen
- 10 1038@nature13906Dokument14 Seiten10 1038@nature13906Diego Benitez RiquelmeNoch keine Bewertungen
- Designs of National Most Cancers InstituteSponsored Medical Trial Enrollment Throughout Black Young People and Also Young Adultsncfdl PDFDokument1 SeiteDesigns of National Most Cancers InstituteSponsored Medical Trial Enrollment Throughout Black Young People and Also Young Adultsncfdl PDFdamagecrab82Noch keine Bewertungen
- WinFiol CommandDokument2 SeitenWinFiol CommandPapitch Do DuneNoch keine Bewertungen
- Wangg 2018Dokument17 SeitenWangg 2018ERAPEWE OFFICIALNoch keine Bewertungen
- 3641 14616 1 PBDokument10 Seiten3641 14616 1 PBEder OliveiraNoch keine Bewertungen
- Monster Synthesis Activity - SFSDokument6 SeitenMonster Synthesis Activity - SFSDiego OFarrillNoch keine Bewertungen
- Jurnal TranskripsiDokument4 SeitenJurnal TranskripsiGisela GloryNoch keine Bewertungen
- Expression of The T-Cell-Specific Adapter Protein in Oral EpitheliumDokument10 SeitenExpression of The T-Cell-Specific Adapter Protein in Oral EpitheliumSuci Dika UtariNoch keine Bewertungen
- Vijaya Sree PPT 2 CDFDDokument44 SeitenVijaya Sree PPT 2 CDFDsai krishnaNoch keine Bewertungen
- 9244-Article Text-64260-3-10-20230708Dokument11 Seiten9244-Article Text-64260-3-10-20230708Benjamin SantOsNoch keine Bewertungen
- Mus Musculus Transformation Related Protein 73 (Trp73), Transcript Variant 3, mRNADokument7 SeitenMus Musculus Transformation Related Protein 73 (Trp73), Transcript Variant 3, mRNAEeeeeeeeeNoch keine Bewertungen
- Protein Kinase C-Dependent Trans-Golgi NetworkDokument9 SeitenProtein Kinase C-Dependent Trans-Golgi NetworkAlix AliNoch keine Bewertungen
- 47 FullDokument7 Seiten47 Fulldrscribd25Noch keine Bewertungen
- SNPDokument60 SeitenSNPRoshan KumarNoch keine Bewertungen
- GCcomputer02 LU EngDokument5 SeitenGCcomputer02 LU EngTiMoNPuNkGiRlNoch keine Bewertungen
- NIHMS563298 Supplement Supplementary - DataDokument5 SeitenNIHMS563298 Supplement Supplementary - DatahNoch keine Bewertungen
- p63 and p73, The Ancestors of p53Dokument15 Seitenp63 and p73, The Ancestors of p53Paata TushurashviliNoch keine Bewertungen
- Dna WsDokument1 SeiteDna WsLovryan Tadena AmilingNoch keine Bewertungen
- Chicken MRNA For IgG H-Chain (HC36) D-J-C - Nucleotide - NCBIDokument2 SeitenChicken MRNA For IgG H-Chain (HC36) D-J-C - Nucleotide - NCBIZulfi Nur Amrina RosyadaNoch keine Bewertungen
- 69th AACC Annual Scientific Meeting Abstract eBookVon Everand69th AACC Annual Scientific Meeting Abstract eBookNoch keine Bewertungen
- PEGFP-N1 Vector InformationDokument3 SeitenPEGFP-N1 Vector InformationNicholas SoNoch keine Bewertungen
- A Transgenic Marc-145 Cell Line of PiggyBac Transposon-DerivedDokument7 SeitenA Transgenic Marc-145 Cell Line of PiggyBac Transposon-DerivedBhaskar RoyNoch keine Bewertungen
- Wild-Type: p63 Irf6Dokument5 SeitenWild-Type: p63 Irf6Nadya PurwantyNoch keine Bewertungen
- 6 CatDokument6 Seiten6 CatbmnlthyukNoch keine Bewertungen
- Poster ApmlDokument1 SeitePoster Apmlkuel1511Noch keine Bewertungen
- Bacterial Gene AnnotationDokument12 SeitenBacterial Gene AnnotationAndrew Steele100% (1)
- Gene, Proteins, and Genetic CodeDokument37 SeitenGene, Proteins, and Genetic CodejuniorNoch keine Bewertungen
- Conserved RNA Secondary Structures in Genomes: PicornaviridaeDokument11 SeitenConserved RNA Secondary Structures in Genomes: Picornaviridaenareman hassanNoch keine Bewertungen
- Pathfast PresepsinDokument6 SeitenPathfast PresepsinWulan Ervinna SimanjuntakNoch keine Bewertungen
- Virus 1Dokument9 SeitenVirus 1Novita RindoNoch keine Bewertungen
- J. Biol. Chem.-2003-Botelho-34259-67Dokument10 SeitenJ. Biol. Chem.-2003-Botelho-34259-67Jose L G FunesNoch keine Bewertungen
- Brain MetabolismDokument12 SeitenBrain MetabolismDr. Kaushal Kishor SharmaNoch keine Bewertungen
- Msa & Sequence DendogramDokument16 SeitenMsa & Sequence DendogramMudit MisraNoch keine Bewertungen
- Biotek StrawberiDokument2 SeitenBiotek StrawberiDesinta PuspitasariNoch keine Bewertungen
- Fasta 1Dokument17 SeitenFasta 1jonathan andre mora quimbayoNoch keine Bewertungen
- App 09Dokument3 SeitenApp 09hvsalgadom6568Noch keine Bewertungen
- An Improved Method of Measurement of Ecg Parameters For Online Medical DiagnosisDokument22 SeitenAn Improved Method of Measurement of Ecg Parameters For Online Medical DiagnosisManikandan VijayakumarNoch keine Bewertungen
- Transferrin: Cdna Chromosomal Localization : Human Characterization andDokument5 SeitenTransferrin: Cdna Chromosomal Localization : Human Characterization andAndrimencitNoch keine Bewertungen
- BCHS317 SU2.5 - Translation - FMDokument55 SeitenBCHS317 SU2.5 - Translation - FMKAGISO BRIAN MOTSHUPHINoch keine Bewertungen
- Molecular Cloning and Functional Analysis of ESGP, An Embryonic Stem Cell and Germ Cell Specific ProteinDokument8 SeitenMolecular Cloning and Functional Analysis of ESGP, An Embryonic Stem Cell and Germ Cell Specific ProteinТимур АбдуллинNoch keine Bewertungen
- DNA Polymorphisms and Human IdentificationDokument44 SeitenDNA Polymorphisms and Human Identificationcracker1983Noch keine Bewertungen
- BioCentralDogmaWorksheet 1Dokument3 SeitenBioCentralDogmaWorksheet 1jaelajeanine326Noch keine Bewertungen
- Genomics of The Severe Isolate of Maize Chlorotic: Dwarf VirusDokument7 SeitenGenomics of The Severe Isolate of Maize Chlorotic: Dwarf VirusBelisa SaitoNoch keine Bewertungen
- PIIS0092867419312115Dokument28 SeitenPIIS0092867419312115LiaaNoch keine Bewertungen
- Sanet - CD 1542982383Dokument204 SeitenSanet - CD 1542982383csodaelmeNoch keine Bewertungen
- KépletgyűjteményDokument3 SeitenKépletgyűjteménycsodaelmeNoch keine Bewertungen
- Bluetooth Network Encapsulation (BNEP) Protocol Test Cases - Rev0.95aDokument41 SeitenBluetooth Network Encapsulation (BNEP) Protocol Test Cases - Rev0.95acsodaelmeNoch keine Bewertungen
- Steen F.wireless NetworkingDokument9 SeitenSteen F.wireless NetworkingcsodaelmeNoch keine Bewertungen
- USB Language Identifiers (LANGIDs) .Rev 1.0.2000Dokument11 SeitenUSB Language Identifiers (LANGIDs) .Rev 1.0.2000csodaelmeNoch keine Bewertungen