Beruflich Dokumente
Kultur Dokumente
Cloning Proyect
Hochgeladen von
Kath rgOriginalbeschreibung:
Copyright
Verfügbare Formate
Dieses Dokument teilen
Dokument teilen oder einbetten
Stufen Sie dieses Dokument als nützlich ein?
Sind diese Inhalte unangemessen?
Dieses Dokument meldenCopyright:
Verfügbare Formate
Cloning Proyect
Hochgeladen von
Kath rgCopyright:
Verfügbare Formate
Cloning proyect 1. What does constitutively active promoter mean?
Why would you want to use a vector with this type of promoter? It is a promoter that is not regulated by any elements allowing continues transcription of its gene. Having a constitutive promoter in a vector will permit the transcription of rscB gene without concerns for gene regulation, which boost RscB expression. 2. Using the protein sequence of RscB in Y. enterocolitica, do a BLAST search to determine if homologs exist in other organisms. If so, what are your top hits (just name 2 or 3)? Rsc B its though to be a surface expose protein. A BLAST search for RscB of Y. enterocolitica showed homology with a hemolysin activation/secretion protein associated with VreARI signaling system and with a supposed lipoproteins in Y. pestis. STRING, a web protein-protein prediction system, predicts interacions with hemolysin and transporter proteins. Hemolysins are proteins that provoke red blood cells lysis. For bacteria, this proteins are important for nutrient acquisition, special iron.
3. Design and outline a strategy to determine the direction that your gene has been cloned into the TOPO vector. Your strategy should include how you would test this as well as an outline of your expected results if your gene is in one direction or the other. Be sure to look up the enzymes that you will be using to do this and outline your protocol with details necessary for you to set up this reaction in a lab.
i) In order to determine the rscB gene direction into the TOPO vector a digestion reaction will be develop. Using the enzymes BbsI and BamHI-HF, digest the DNA. The product of this reaction will be two DNA sequence. If the vector is in the forward direction, one band of ~4.6kbp and one band of ~309bp would be seen when a 2% agarose gel is run (Figure 1). In the other hand, if the rscB gene is the reverse direction, one band of ~4.2kbp and one band of ~719bp would be reflected in the agarose gel after the electrophoresis (Figure 2). In addition, positive and negative controls are highly recommended. ii) Rx set up: (1) Digestion Rx: 36 uL water, 5.0uL BSA, 5.0 uL NE Buffer 1, 1.5 uL DNA (500ng of DNA to observe at ~30-80ng of DNA in the smaller possible band), 2.0 uL Bbs 1 (10 units), 0.5 uL BamHI-HF (10 units). (2) Negative control: 38.5 uL water, 5.0uL BSA, 5.0 uL NE Buffer 1, 1.5 uL DNA. (3) Positive control for Bbs 1: 36.5 uL water, 5.0uL BSA, 5.0 uL NE Buffer 1, 1.5 uL DNA, 2.0 uL Bbs 1. (4) Positive control for BamHI-HF: 38 uL water, 5.0uL BSA, 5.0 uL NE Buffer 1, 1.5 uL DNA, 0.5 uL BamHI-HF. iii) Run the reaction for at least 1hr @ 37 C.
iv) Run a 2% agarose gel electrophoresis. Load 5uL with 1X loading dye (1uL from 6X) from each reaction into a well. v) Expected results Figure 1
Figure 2
4. Design and outline a strategy to move your gene into the TOPO vector. Your strategy should include the restriction enzymes that you would use to move your rscB gene into the pWKS130 vector (taking into account the direction it is in the TOPO vector), as well as a strategy to verify you have cloned it in the proper direction in the pWKS 130 vector and your expected results. Be sure to look up the enzymes that you will be using to do this and outline your protocol with details necessary for you to set up this reaction in a lab.
i) Depending on the results from agarose gel, you will pick to digest for the forward or reverse direction in order to have the correct sequence of the start codon next to the promoter in the pWKS130 vector. (1) TOPO + insert digestion: (a) Digestion for forward: 1hr @ 37 C 1. BamHI-HF: 0.5 uL 2. Xbal: 0.5 uL 3. DNA (TOPO + insert): 10 uL (10000ng ~1800ng of Insert) 4. 1X Ne Buffer 4: 5 uL 5. 1X BSA: 5 uL 6. dd water: 29 uL (b) or Digestion for reverse: 1. Xho 1: 0.5 uL 2. BamHI-HF: 0.5 uL 3. DNA (TOPO + insert): 10 uL (10000ng ~1800ng of Insert) 4. 1X Ne Buffer 4: 5 uL 5. 1X BSA: 5 uL 6. dd water: 29 uL (2) pWKS130 vector digestion: 1hr @ 37 C (a) Digestion for forward: 1. BamHI-HF: 0.5 uL 2. Xbal: 0.5 uL 3. DNA: 4uL for 1000ng 4. 1X Ne Buffer 4: 5 uL 5. 1X BSA: 5 uL 6. dd water: 35 uL (b) or Digestion for reverse: 1. Xho 1: 0.5 uL 2. BamHI-HF: 0.5 uL 3. DNA: 4 uL for 1000ng 4. 1X Ne Buffer 4: 5 uL 5. 1X BSA: 5 uL 6. dd water: 35 uL ii) Run 5 uL of the digestion product in a 5% agarose gel electrophoresis. This will run ~180ng of the insert DNA in the gel. To dilute the DNA add 20 uL of dd water. iii) Isolate the correct bands. ~900bp for insert iv) Purify the DNA band on the gel following the QUIK gen purification protocol. v) Quantify the DNA concentration.
vi) Ligation: ligate the insert into the pWKS130 vector in a 3:1 ratio. (1) Add 16 ng of vector to 50 ng of insert, and adjust the volume for 10 uL with ddwater. (2) Add 10 l of 2X Quick Ligation Buffer and mix (3) Add 1l of Quick T4 DNA Ligase and mix (4) Centrifuge briefly and incubate at room temperature (25C) for 5 minutes. (5) The ligation product is ready to use. Store for until use on ice.
Das könnte Ihnen auch gefallen
- Users Guide For Phosphoprotein ReagentsDokument9 SeitenUsers Guide For Phosphoprotein Reagentsthumita kumiNoch keine Bewertungen
- Nar00406 0009Dokument10 SeitenNar00406 0009Adulrazaq AtawiNoch keine Bewertungen
- Early Sporulation GeneDokument5 SeitenEarly Sporulation GeneStephen G. SabinayNoch keine Bewertungen
- Lab Report 1Dokument14 SeitenLab Report 1esn_kNoch keine Bewertungen
- Jbacter00209 0028Dokument9 SeitenJbacter00209 0028edal_108Noch keine Bewertungen
- Immunobiology - Lab Report 2Dokument8 SeitenImmunobiology - Lab Report 2xjakeknockout0% (1)
- Gateway Technology: M.Phil. Biotechnology CBM, University of SwatDokument9 SeitenGateway Technology: M.Phil. Biotechnology CBM, University of SwatGul AfshaNoch keine Bewertungen
- New Tool For Metabolic Pathway Engineering in Escherichia Coli: One-Step Method To Modulate Expression of Chromosomal GenesDokument5 SeitenNew Tool For Metabolic Pathway Engineering in Escherichia Coli: One-Step Method To Modulate Expression of Chromosomal Genesaib_himaniNoch keine Bewertungen
- 5 Complementation of Apolipoprotein B mRNA Editing by Human Liver Accompanied by Secretion of Apolipoprotein B48 J. Biol. Chem.-1994-Giannoni-5932-6Dokument5 Seiten5 Complementation of Apolipoprotein B mRNA Editing by Human Liver Accompanied by Secretion of Apolipoprotein B48 J. Biol. Chem.-1994-Giannoni-5932-6Federico GiannoniNoch keine Bewertungen
- tmpB2BC TMPDokument6 SeitentmpB2BC TMPFrontiersNoch keine Bewertungen
- V. CholeraeDokument6 SeitenV. CholeraeEstefani BlancasNoch keine Bewertungen
- BFP To GFPDokument11 SeitenBFP To GFPKeri Gobin SamarooNoch keine Bewertungen
- MMP-1 Practical Handout v1.0Dokument5 SeitenMMP-1 Practical Handout v1.0Maisha JashimNoch keine Bewertungen
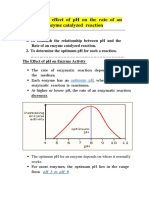
- Effect of PH On Enzyme ActivityDokument12 SeitenEffect of PH On Enzyme ActivityAb AbNoch keine Bewertungen
- Chemistry and Physics of Lipids: A A B A ADokument6 SeitenChemistry and Physics of Lipids: A A B A Aannisya bubblesNoch keine Bewertungen
- Molecular Cell Biology Practice QuestionsDokument9 SeitenMolecular Cell Biology Practice Questionssahana2791100% (1)
- TMP AF40Dokument4 SeitenTMP AF40FrontiersNoch keine Bewertungen
- Reece-Hoyes Gateway Recombinational CloningDokument7 SeitenReece-Hoyes Gateway Recombinational CloningJosue VillotaNoch keine Bewertungen
- Lab ReportDokument7 SeitenLab ReportAlliedschool DefencecampusNoch keine Bewertungen
- (A) Identification of A New LAP Biosynthetic Gene ClusterDokument5 Seiten(A) Identification of A New LAP Biosynthetic Gene ClusterPappu HSD VenkateshNoch keine Bewertungen
- PX330 Cloning ProtocolDokument4 SeitenPX330 Cloning ProtocolSubhajit DuttaNoch keine Bewertungen
- Zac 4651Dokument6 SeitenZac 4651bondester4Noch keine Bewertungen
- Drosophila Midgut, and Developing Chick Retina, Suggesting That It Is Likely Useful For Most OrganismsDokument23 SeitenDrosophila Midgut, and Developing Chick Retina, Suggesting That It Is Likely Useful For Most Organismsk.sachinNoch keine Bewertungen
- Bio E1a 2016 Review Outline C 4Dokument7 SeitenBio E1a 2016 Review Outline C 4FaisalNoch keine Bewertungen
- EstrogenicDokument2 SeitenEstrogenici_ghanemNoch keine Bewertungen
- NIHMS687333 Supplement 2Dokument28 SeitenNIHMS687333 Supplement 2zak marshallNoch keine Bewertungen
- Dissertation Talk7Dokument39 SeitenDissertation Talk7Chris McGeeNoch keine Bewertungen
- 16S Ribosomal DNA For: Amplification Phylogenetic StudyDokument7 Seiten16S Ribosomal DNA For: Amplification Phylogenetic StudyolamicroNoch keine Bewertungen
- Weisburg Et Al 1991 16s Ribosomal Dna Amplification For Phylogenetic StudyDokument7 SeitenWeisburg Et Al 1991 16s Ribosomal Dna Amplification For Phylogenetic Studyteacher.mireyaNoch keine Bewertungen
- Protein Membrane Overlay AssayDokument4 SeitenProtein Membrane Overlay AssayZain BaderNoch keine Bewertungen
- Pnas 1613199114 SappDokument37 SeitenPnas 1613199114 SappLavanya MittalNoch keine Bewertungen
- Practical 3. Analyzing DNA FragmentsDokument9 SeitenPractical 3. Analyzing DNA FragmentsLloaana 12Noch keine Bewertungen
- 7.013 Recitation 3 - Spring 2015: Summary of Lecture 4Dokument5 Seiten7.013 Recitation 3 - Spring 2015: Summary of Lecture 4CeriseNoch keine Bewertungen
- Research Project Full - OdtDokument13 SeitenResearch Project Full - OdtAnonymous 7RSTHfJuGNNoch keine Bewertungen
- Chem 43 - Worksheet3 - NADokument2 SeitenChem 43 - Worksheet3 - NAFock StudentNoch keine Bewertungen
- Journal of Pharmacological and Toxicological Methods: How ToDokument11 SeitenJournal of Pharmacological and Toxicological Methods: How ToMichelleNoch keine Bewertungen
- Southern Blot Analysis of Plasmids pRIT4501 and pRIT4502: Experiment 18Dokument10 SeitenSouthern Blot Analysis of Plasmids pRIT4501 and pRIT4502: Experiment 18Nur SyamimiNoch keine Bewertungen
- Intracellular Hepatitis C Virus RNA-dependent RNA Polymerase ActivityDokument4 SeitenIntracellular Hepatitis C Virus RNA-dependent RNA Polymerase ActivitybmnlthyukNoch keine Bewertungen
- 7.016 Recitation 3 - Fall 2018: (Note: The Recitation Summary Should NOT Be Regarded As The Substitute For Lectures)Dokument6 Seiten7.016 Recitation 3 - Fall 2018: (Note: The Recitation Summary Should NOT Be Regarded As The Substitute For Lectures)Manish SarkarNoch keine Bewertungen
- Lyngstadaas 1999Dokument9 SeitenLyngstadaas 1999Fede ZannierNoch keine Bewertungen
- Dorseuil Et Al 1992 - Inhibition of Superoxide Production in B Lymphocytes by Rac Antisense OligonucleotidesDokument3 SeitenDorseuil Et Al 1992 - Inhibition of Superoxide Production in B Lymphocytes by Rac Antisense OligonucleotidesHernestoNoch keine Bewertungen
- Forsberg Et Al 1994 Use of Transcriptional Fusions To Monitor Gene Expression A Cautionary TaleDokument5 SeitenForsberg Et Al 1994 Use of Transcriptional Fusions To Monitor Gene Expression A Cautionary TaleKunal KumarNoch keine Bewertungen
- Exp3 FinalDokument14 SeitenExp3 Finalapi-253261856Noch keine Bewertungen
- Rapid and Efficient Cosmid Cloning: D.Ish-Horowicz and J.F.BurkeDokument10 SeitenRapid and Efficient Cosmid Cloning: D.Ish-Horowicz and J.F.BurkerezqNoch keine Bewertungen
- Bio L 100 Bacterial Trans LabDokument11 SeitenBio L 100 Bacterial Trans LabRohit ReddyNoch keine Bewertungen
- Deteksi Molekuler 2Dokument10 SeitenDeteksi Molekuler 2CHAYRA STUDIONoch keine Bewertungen
- Zhang2018transcriptome Sequencing RNA-SeqDokument13 SeitenZhang2018transcriptome Sequencing RNA-SeqKito TongHuiNoch keine Bewertungen
- Signal Transduction Methods - LabDokument11 SeitenSignal Transduction Methods - Lab42030570Noch keine Bewertungen
- Pnas 1402658111 SappDokument25 SeitenPnas 1402658111 SappAmín MoraNoch keine Bewertungen
- Identification of Conformational Antigenic Epitopes and Dominant Amino Acids of Buffalo β-LactoglobulinDokument9 SeitenIdentification of Conformational Antigenic Epitopes and Dominant Amino Acids of Buffalo β-LactoglobulinJhudit CamachoNoch keine Bewertungen
- Ekspresi Protein RekombinanDokument35 SeitenEkspresi Protein RekombinanFenny rahmadhanyNoch keine Bewertungen
- Chapter 7Dokument5 SeitenChapter 7Leonita Swandjaja100% (1)
- Involvement of The Rab27 Binding Protein Slac2C/Myrip in Insulin ExocytosisDokument11 SeitenInvolvement of The Rab27 Binding Protein Slac2C/Myrip in Insulin ExocytosisJoseGonzalezNoch keine Bewertungen
- Week 1 RNA ExtractionDokument11 SeitenWeek 1 RNA ExtractionArjun BujjiNoch keine Bewertungen
- Molcellb00093 0072Dokument8 SeitenMolcellb00093 0072Milan PetrikNoch keine Bewertungen
- DNA Repair Protein Involved in Heart and Blood DevelopmentDokument12 SeitenDNA Repair Protein Involved in Heart and Blood DevelopmentSol Jumaide WerbleNoch keine Bewertungen
- Bte 204 Assignment: Name: Sadman Rashid Abir ID: 20136004 SEC: 01Dokument7 SeitenBte 204 Assignment: Name: Sadman Rashid Abir ID: 20136004 SEC: 01Sadman RashidNoch keine Bewertungen
- Laporan Amali Jib 322 (841009-14-5879 - JP957115)Dokument37 SeitenLaporan Amali Jib 322 (841009-14-5879 - JP957115)Jayanthi Loganathan100% (1)
- AssstDokument5 SeitenAssstDiegoNoch keine Bewertungen
- Analysis of Protein Post-Translational Modifications by Mass SpectrometryVon EverandAnalysis of Protein Post-Translational Modifications by Mass SpectrometryNoch keine Bewertungen
- Vol 30 Profiles - of - Drug - Substances, - E (BookSee - Org) Vol 30Dokument329 SeitenVol 30 Profiles - of - Drug - Substances, - E (BookSee - Org) Vol 30Gabriel Bianco100% (1)
- Learning Activity 4 Evidence: My View On Colombia: 1. Write A Composition Describing What The LifestyleDokument2 SeitenLearning Activity 4 Evidence: My View On Colombia: 1. Write A Composition Describing What The LifestyleYenifer PatiñoNoch keine Bewertungen
- PET SyllabusDokument2 SeitenPET SyllabusAureabeNoch keine Bewertungen
- Character Strengths and Virtues A Handbook of Classification MindmapDokument1 SeiteCharacter Strengths and Virtues A Handbook of Classification MindmapJorge Luis Villacís NietoNoch keine Bewertungen
- Classroom and Lab Area - Job Roles Wise2Dokument117 SeitenClassroom and Lab Area - Job Roles Wise2Param ShikshaNoch keine Bewertungen
- 2022 CA Security Assessment and Authorization StandardDokument25 Seiten2022 CA Security Assessment and Authorization StandardDonaldNoch keine Bewertungen
- Young Children's Use of Digital Media and The Corresponding Parental MediationDokument15 SeitenYoung Children's Use of Digital Media and The Corresponding Parental MediationFernan EnadNoch keine Bewertungen
- Jean Watson TheoryDokument20 SeitenJean Watson TheorySandara Myrrh CalumbaNoch keine Bewertungen
- Mr./Ms. Anshu Yadav Dear Anshu,: DateDokument3 SeitenMr./Ms. Anshu Yadav Dear Anshu,: DateAnshu YadavNoch keine Bewertungen
- MotherDokument11 SeitenMotherMuhammad Lucky AlfaridzyNoch keine Bewertungen
- Common Rail System (CRS) SERVICE MANUAL: Operation: YD2K3 EngineDokument44 SeitenCommon Rail System (CRS) SERVICE MANUAL: Operation: YD2K3 EngineAntony ColonnaNoch keine Bewertungen
- Summary of Cutting Data For Plain Surface: DegrosareDokument4 SeitenSummary of Cutting Data For Plain Surface: DegrosareAndrei MihaiNoch keine Bewertungen
- Activity Sheet 3 in Science ViDokument4 SeitenActivity Sheet 3 in Science ViJohn Cyrel MondejarNoch keine Bewertungen
- PENC N BONDIDokument5 SeitenPENC N BONDIYLND100% (1)
- C1 - C7 General Requirements For Equipment ErectionDokument24 SeitenC1 - C7 General Requirements For Equipment ErectionephNoch keine Bewertungen
- Enabling Works SpecificationDokument290 SeitenEnabling Works SpecificationMAJ19800% (2)
- People in OrganisationsDokument8 SeitenPeople in OrganisationsBritney valladares100% (1)
- Traffic Laws Rules and Regulations and Other Related Statutes.Dokument23 SeitenTraffic Laws Rules and Regulations and Other Related Statutes.Leo AnchetaNoch keine Bewertungen
- Steca TR 0201 Instruction enDokument40 SeitenSteca TR 0201 Instruction endaviko313Noch keine Bewertungen
- Final - Urban and Transportation Engineering - PPT Group 1Dokument19 SeitenFinal - Urban and Transportation Engineering - PPT Group 1JayChristian QuimsonNoch keine Bewertungen
- TOXICITY of FLOURIDESDokument45 SeitenTOXICITY of FLOURIDESNavneet KaurNoch keine Bewertungen
- Installation User Instruction (KMC Corporation)Dokument33 SeitenInstallation User Instruction (KMC Corporation)Dan StroescuNoch keine Bewertungen
- W - Paul Smith, William C. Compton and W - Beryl WestDokument5 SeitenW - Paul Smith, William C. Compton and W - Beryl Westefarfan26Noch keine Bewertungen
- AN5276 Application Note: Antenna Design For ST25R3916Dokument43 SeitenAN5276 Application Note: Antenna Design For ST25R3916RafaelNoch keine Bewertungen
- Á641Ñ Completeness of Solution: 310 Á631ñ Color and Achromicity / Physical Tests USP 37Dokument2 SeitenÁ641Ñ Completeness of Solution: 310 Á631ñ Color and Achromicity / Physical Tests USP 37David SRNoch keine Bewertungen
- Pharmacist Workup of Drug Therapy in Pharmaceutical Care: Problem Oriented Pharmacist RecordDokument20 SeitenPharmacist Workup of Drug Therapy in Pharmaceutical Care: Problem Oriented Pharmacist RecordNurwahidah Moh WahiNoch keine Bewertungen
- 1HDH 118 041 en - Rev. IDokument53 Seiten1HDH 118 041 en - Rev. IKhắc Đồng Trần100% (1)
- Xtralift NBS Specification MAY21Dokument4 SeitenXtralift NBS Specification MAY21Ahmed AbdelftahNoch keine Bewertungen
- AS 2122 HK1 DecuongHKI ANH7Dokument19 SeitenAS 2122 HK1 DecuongHKI ANH7Nguyễn Lê KhánhNoch keine Bewertungen
- Pile Hammer Delmag d12-42Dokument1 SeitePile Hammer Delmag d12-42Jorge Rosero QuevedoNoch keine Bewertungen